Automatic Pancreas Segmentation using A Novel Modified Semantic Deep Learning Bottom-Up Approach
DOI:
https://doi.org/10.18201/ijisae.2022.272Keywords:
Deep Learning, BFscore, Dice Coefficient, Visual Geometry Group(VGG)Abstract
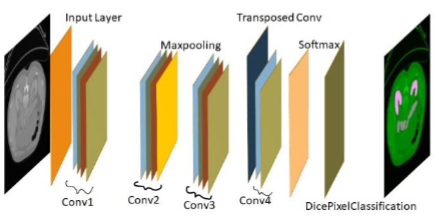
Sharpe and smooth pancreas segmentation is crucial and arduous problem in medical image analysis and investigation. A semantic deep learning bottom- up approach is most popular and efficient method used for pancreas seg- mentation with smooth and sharp result. Automatic pancreas segmentation process is performed through semantic segmentation for abdominal computed tomography(CT) clinical images. A novel semantic segmentation is applied for acute pancreas segmentation with different angle of CT images. In novel modified semantic approach 12 layers are used. The proposed model is exe- cuted on a dataset of 80 patient single phase CT image. For training purpose 699 images and testing purpose 150 images are taken from dataset with dif- ferent angle. Proposed approach is used for many organ segmentation from CT scans clinical images with high accuracy. Computation time period is reduced as related to the state-of-art. Validation accuracy is a 69.29% and loss values varies 1 to 0 only. Bfscore,Dice Coefficient, Jaccard Coefficient are used to calculate similarity index values between test image and expected output image only. The proposed approach achieved a dice similarity index score upto 81±7.43%. Class balancing process is executed with the help of class weight and data augmentation. In novel modified semantic segmen- tation, max-pooling layer, RELU layer, softmax layer, transposed conv2d layer and dicePixelClassification layer are used. DicePixelClassification is newly introduced and incorporated in novel method for improved results. VGG-16,VGG-19 and RSnet-18 deep learning models are used for pancreas segmentation.
Downloads
References
Paithane P.M., Kakarwal S.N., Kurmude D.V. ,"Automatic Seeded Region Growing with Level Set Technique Used for Segmentation of Pancreas." Proceedings of the 12th International Conference on Soft Computing and Pattern Recognition (SoCPaR 2020). SoCPaR 2020. Advances in Intelligent Systems and Computing, vol 1383. Springer, Cham.(2021)
Amal Farag, Le Lu, Holger R. Roth, Jiamin Liu, Evrim Turkbey, and Ronald M. Summers,"A Bottom-Up Approach for Pancreas Segmentation Using Cascaded Superpixels and (Deep) Image Patch Labeling",IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 26, NO. 1, JANUARY 2017,Digital Object Identifier 10.1109/TIP.2016.2624198
Paithane, P.M., Kakarwal, S.N., Kurmude, D.V.," Top-down method used for pancreas segmentation." International Journal Innovation Exploring Engineering. (IJITEE) 9(3), 1790–1793 (2020). ISSN 2278-3075
Harshita Sharmaa, Norman Zerbeb, Iris Klempertb, Olaf Hellwicha, Peter Hufnagl,"Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology", Computerized Medical Imaging andGraphics,(2017), http://dx.doi.org/10.1016/j.compmedimag.2017.06.001
J. Long, E. Shelhamer, and T. Darrell, “Fully convolutional networks for semantic segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 3431–3440.
Geert Litjens , Thijs Kooi , Babak Ehteshami Bejnordi , Arnaud Arindra Adiyoso Setio , Francesco Ciompi, Mohsen Ghafoorian, JeroenA.W.M. van der Laak, Bram van Ginneken, Clara I. Sánchez,"A survey on deep learning in medical image analysis",Medical Image Analysis,42,2017,pp. 60–88
Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Armbruster M, Hofmann F, D’Anastasi M,"Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields". International Conference on Medical Image Computing
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: https://doi.org/10.1007/s10278-013-9622-7
Glorot, Xavier, and Yoshua Bengio. "Understanding the Difficulty of Training Deep Feedforward Neural Networks." In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, 249–356. Sardinia, Italy: AISTATS, 2010.
Shervin Minaee,Yuri Boykov,Fatih Porikli,Antonio Plaza,Nasser Kehtarnavaz, and Demetri Terzopoulos,"Image Segmentation Using Deep Learning:A Survey",TRANSACTIONS ON PATTERN ANALYSIS AND MACHINE INTELLIGENCE, 2021
Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, Davidson BR, Pereira SP, Clarkson MJ, Barratt DC,"Towards image-guided pancreas and biliary endoscopy: automatic multiorgan segmentation on abdominal CT with dense dilated networks." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2017, pp 728–736
Mohammad Hesam Hesamian1,Wenjing Jia1,Xiangjian He1,Paul Kennedy,"Deep Learning Techniques for Medical Image Segmentation:Achievements and Challenges",Journal of Digital Imaging,Springer,2019
Krizhevsky, A., I. Sutskever, and G. E. Hinton. "ImageNet Classification with Deep Convolutional Neural Networks". Advances in Neural Information Processing Systems. Vol 25, 2012.
Nagi, J., F. Ducatelle, G. A. Di Caro, D. Ciresan, U. Meier, A. Giusti, F. Nagi, J. Schmidhuber, L. M. Gambardella. ”Max-Pooling Convolutional Neural Networks for Vision-based Hand Gesture Recognition”. IEEE International Conference on Signal and Image Processing Applications (ICSIPA2011), 2011
He, Kaiming, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. "Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification." In Proceedings of the 2015 IEEE International Conference on Computer Vision, 1026–1034. Washington, DC: IEEE Computer Vision Society, 2015.
Sarfaraz Hussein , Pujan Kandel, Candice W. Bolan, Michael B. Wallace, Ulas Bagci,"Lung and Pancreatic Tumor Characterization in the Deep Learning Era: Novel Supervised and Unsupervised Learning Approaches.",IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 38, NO. 8, AUGUST 2019,pp.1777-1787
Guanjin Wang , Ta Zhou , Kup-Sze Choi , and Jie Lu ,"A Deep-Ensemble-Level-Based Interpretable Takagi–Sugeno–Kang Fuzzy Classifier for Imbalanced Data." IEEE TRANSACTIONS ON CYBERNETICS,2168-2267, 2020
Bishop, C. M.." Pattern Recognition and Machine Learning". Springer, New York, NY, 2006.
Csurka, G., D. Larlus, and F. Perronnin. "What is a good evaluation measure for semantic segmentation?" Proceedings of the British Machine Vision Conference, 2013, pp. 32.1–32.11.
Crum, William R., Oscar Camara, and Derek LG Hill. "Generalized overlap measures for evaluation and validation in medical image analysis." IEEE Transactions on Medical Imaging. 25.11, 2006, pp. 1451–1461
Robin Wolz, Chengwen Chu, Kazunari Misawa, Michitaka Fujiwara, Kensaku Mori, and Daniel Rueckert,"Automated Abdominal Multi-Organ Segmentation With Subject-Specific Atlas Generation",IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 32, NO. 9, SEPTEMBER 2013
Ozan Oktay1,Jo Schlemper1, Loic Le Folgoc1, Matthew Lee, Mattias Heinrich,Kazunari Misawa, Kensaku Mori, Steven McDonagh, Nils Y Hammerla,Bernhard Kainz1, Ben Glocker, and Daniel Rueckert1,"Attention U-Net:Learning Where to Look for the Pancreas",1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands.
Paithane, P.M., Kakarwal, S.N.: Automatic determination number of cluster for multi kernel NMKFCM algorithm on image segmentation. In: Intelligent System Design and Applications, pp. 80–89. Springer, Cham, (2018).
Holger R. Roth, Amal Farag, Evrim B. Turkbey, Le Lu, Jiamin Liu, and Ronald M. Summers. (2016). Data From Pancreas-CT. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.tNB1kqBU
Roth HR, Lu L, Farag A, Shin H-C, Liu J, Turkbey EB, Summers RM. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. N. Navab et al. (Eds.): MICCAI 2015, Part I, LNCS 9349, pp. 556–564, 2015.
Paithane, P.M., Kinariwal, S.A.: Automatic determination number of cluster for NMKFC-means algorithm on image segmentation. IOSR-JCE 17(1), 12–19 . Ver 2.(2015).
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 Pradip Mukundrao Paithane, Dr.S.N. Kakarwal

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.









