A Smartphone Application for Skin Lesion Detection and Classification with Deep Learning Algorithms
Keywords:
Application development, Customized model, Deep models, Skin lesion classification, Tensor Flow Lite (TFL), Validation accuracyAbstract
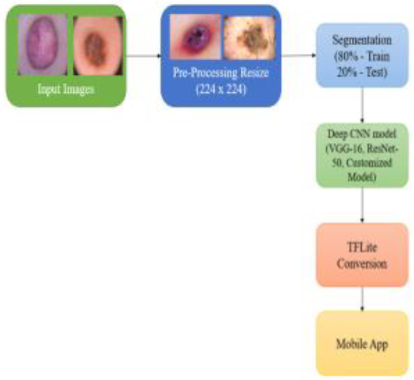
The Skin Lesion (SL) classification has recently received a lot of attention. Because of the significant resemblance between these skin lesions, physicians spend a lot of time analyzing them. A Deep Learning (DL) based automated categorization system can help clinicians recognize the type of SL and improve the patient's health. In this research, DL approaches such as VGG-16, ResNet-50 and customized model are employed to detect the SL using a smartphone application. These models are trained on the SL classification dataset from the International Skin Imaging Collaboration (ISIC) 2019. The customized model over fits the other two models with a validation accuracy of 86.21%, whereas the validation accuracy of VGG-16 and ResNet-50 is 85.15% and 84.82%, respectively. Physicians will save time and have a higher precision rate in the automatic classification of SL utilizing DL.
Downloads
References
WHO-World Health Organization. “How common is skin cancer?” 2019 [Online]. Available: https://www.who.int/uv/faq/skincancer/en/ index1.html
WHO - World Health Organization. “Health effects of UV radiation”. 2019 [Online]. Available: https://www.who.int/uv/health/uv health2/en/index1.html
Jat, N. C., and C. . Kumar. “Design Assessment and Simulation of PCA Based Image Difference Detection and Segmentation for Satellite Images Using Machine Learning”. International Journal on Recent and Innovation Trends in Computing and Communication, vol. 10, no. 3, Apr. 2022, pp. 01-11, doi:10.17762/ijritcc.v10i3.5520.
World Health Organization. “Radiation: Ultraviolet (UV) Radiation and Skin Cancer”. Available online: https://www.who.int/ news-room/questions-and-answers/item/radiation-ultraviolet-(uv)-radiation-andskincancer#:~{}:text=Currently%2C%20between%202%20and%203,skin%20cancer%20in%20their%20lifetime (accessed on 19 October 2021).
Zhang, N., Cai, Y.X., Wang, Y.Y., Tian, Y.T., Wang, X.L. and Badami, B., “Skin cancer diagnosis based on optimized convolutional neural network. Artificial intelligence in medicine”, 2020, 102, pp.101756.
Deepak Mathur, N. K. V. . (2022). Analysis & Prediction of Road Accident Data for NH-19/44. International Journal on Recent Technologies in Mechanical and Electrical Engineering, 9(2), 13–33. https://doi.org/10.17762/ijrmee.v9i2.366
Brinker, T.J., Hekler, A., Enk, A.H., Berking, C., Haferkamp, S., Hauschild, A., Weichenthal, M., Klode, J., Schadendorf, D., Holland-Letz, T. and von Kalle, C.,. “Deep neural networks are superior to dermatologists in melanoma image classification”. European Journal of Cancer, 2019, 119, pp.11-17.
Esteva, A., Kuprel, B., Novoa, R.A., Ko, J., Swetter, S.M., Blau, H.M. and Thrun, S., “Dermatologist-level classification of skin cancer with deep neural networks. Nature”, 2019, 542(7639), pp.115-118.
Hekler, A., Utikal, J.S., Enk, A.H., Hauschild, A., Weichenthal, M., Maron, R.C., Berking, C., Haferkamp, S., Klode, J., Schadendorf, D. and Schilling, B., “Superior skin cancer classification by the combination of human and artificial intelligence. European Journal of Cancer”, 2019, 120, pp.114-121.
Krishnaveni, S. ., A. . Lakkireddy, S. . Vasavi, and A. . Gokhale. “Multi-Objective Virtual Machine Placement Using Order Exchange and Migration Ant Colony System Algorithm”. International Journal on Recent and Innovation Trends in Computing and Communication, vol. 10, no. 6, June 2022, pp. 01-09, doi:10.17762/ijritcc.v10i6.5618.
Feng, H., Berk-Krauss, J., Feng, P.W. and Stein, J.A., “Comparison of dermatologist density between urban and rural counties in the United States”. JAMA dermatology, 2018, 154(11), pp.1265-1271.
O'Dea, S. (2022). “Forecast number of mobile users worldwide 2020-2025”. July 21 2021 Available : https://www.statista.com/statistics/218984/number-of-global-mobile-users-since-2010/
Deloitte (2021) Deloitte report [Online]. Available: (https://www.business-standard.com/article/current-affairs/india-to-have-1-billion-smartphone-users-by-2026-deloitte-report-122022200996_1.html
Harangi, B., “Skin lesion classification with ensembles of deep convolutional neural networks”. Journal of biomedical informatics, 2018, 86, pp.25-32.
Brinker, T.J., Hekler, A., Enk, A.H., Berking, C., Haferkamp, S., Hauschild, A., Weichenthal, M., Klode, J., Schadendorf, D., Holland-Letz, T. and von Kalle, C., “Deep neural networks are superior to dermatologists in melanoma image classification. European Journal of Cancer, 2017,19, pp.11-17.
Arik, A., Gölcük, M. and Karslıgil, E.M “ Deep learning based skin cancer diagnosis”. Signal Processing and Communications Applications Conference (SIU), 2017, (pp. 1-4). IEEE.
Demir, A., Yilmaz, F. and Kose, O., 2019, October. “Early detection of skin cancer using deep learning architectures: resnet-101 and inception-v3”. medical technologies congress (TIPTEKNO) 2019, (pp. 1-4). IEEE.
Aggarwal, A., Das, N. and Sreedevi, I., “Attention-guided deep convolutional neural networks for skin cancer classification” Ninth International Conference on Image Processing Theory, Tools and Applications (IPTA) (pp. 1-6). 2019, IEEE.
Chaudhary, D. S. . (2022). Analysis of Concept of Big Data Process, Strategies, Adoption and Implementation. International Journal on Future Revolution in Computer Science &Amp; Communication Engineering, 8(1), 05–08. https://doi.org/10.17762/ijfrcsce.v8i1.2065
Phillips, K., Fosu, O. and Jouny, I., “Mobile melanoma detection application for android smart phones”. In 41st Annual Northeast Biomedical Engineering Conference (NEBEC), 2015 (pp. 1-2). IEEE.
Castro, P.B., Krohling, B., Pacheco, A.G. and Krohling, R.A., “An app to detect melanoma using deep learning: An approach to handle imbalanced data based on evolutionary algorithms”. In International Joint Conference on Neural Networks (IJCNN), 2020, (pp. 1-6). IEEE.
Simonyan, K. and Zisserman, A., “Very deep convolutional networks for large-scale image recognition.” arXiv preprint arXiv:2020, 1409.1556.
Patil, V. N., & Ingle, D. R. (2022). A Novel Approach for ABO Blood Group Prediction using Fingerprint through Optimized Convolutional Neural Network. International Journal of Intelligent Systems and Applications in Engineering, 10(1), 60–68. https://doi.org/10.18201/ijisae.2022.268
He, K., Zhang, X., Ren, S. and Sun, J., “Deep residual learning for image recognition”. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, (pp. 770-778).
Tan, M.; Le, Q. “Efficient net: Rethinking model scaling for convolutional neural network”s.arXiv 2019, arXiv:1905.11946
Hoang, L.; Lee, S.-H.; Lee, E.-J.; Kwon, K.-R. “Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare. ”Appl. Sci. 2022, 12, 2677. https://doi.org/10.3390/app12052677
Anusha, D. J. ., R. . Anandan, and P. V. . Krishna. “Modified Context Aware Middleware Architecture for Precision Agriculture”. International Journal on Recent and Innovation Trends in Computing and Communication, vol. 10, no. 7, July 2022, pp. 112-20, doi:10.17762/ijritcc.v10i7.5635.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





