A Deep Convolutional Extreme Machine Learning Classification Method To Detect Bone Cancer From Histopathological Images
Keywords:
bone cancer, histopathology, classification, machine learning, ensemble classifierAbstract
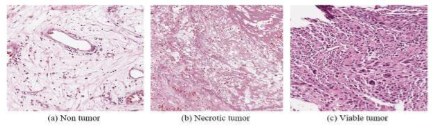
The histopathology process remains traditional in nature. Additionally, the physical process of pathologists could deal with solely restricted subjects because of extended phases. This physical process might misguide the physicians when there remain mass subjects to diagnose because of limited time and the nature of complex illnesses such as bone cancer. Addressing this study in digital histopathology remains significant by evolving computer-aided instruments for detection. Bone framework intricacy remains the chief cause to be a gray research field. Comprehension and investigation of the disparate extent of bone anatomy would serve the requirement for building study in automation. To classify the density tumor Computer-aided diagnosis systems have been developed, having as a major challenge to define the features that better represent the images to classify. To overcome the problem, this paper aims to develop a Convolutional Extreme Learning Machine (DC-ELM) algorithm for the assessment of cancer type based on analyzing histopathology images. A fusion of five important classifiers is chosen for our method. In the framework proposed, only by utilizing the karhunenloeve extraction technique, we extract such and such features of the image and, thus, for differentiation of healthy and unhealthy bone regions.
These extracted features are fed to machine learning architecture with the prediction for achieving better accuracy. As a result, the suggested DC-ELM algorithm achieves 97.27% accuracy.
Downloads
References
Nisthula P andYadhu.R.B, “A Novel Method to Detect Bone Cancer using Image Fusion and Edge Detection”, Proceedings of Engineering and Computer Science, vol.3, no.2, pp.2012- 2018, 2013.
Hubert H. Chuang, Beth A. Chasen, and TinsuPan, “Bone Cancer Detection from MRI Scan Imagery Using Mean Pixel Intensity”, Proceedings of Advanced Research in Computer and Software Engineering, vol.1, no.5, pp.1-7, 2013.
Balaji, G. N., T. S. Subashini, and A. Manikandarajan. "Automatic Classification of Anterio-Posterior and Lateral Views of Leg X-rays”, International Research Journal of Engineering and Technology (IRJET), Vol.2, no.9, pp.968-974, 2015.
Tume-Bruce, B. A. A. ., A. . Delgado, and E. L. . Huamaní. “Implementation of a Web System for the Improvement in Sales and in the Application of Digital Marketing in the Company Selcom”. International Journal on Recent and Innovation Trends in Computing and Communication, vol. 10, no. 5, May 2022, pp. 48-59, doi:10.17762/ijritcc.v10i5.5553.
Balaji, G. N., T. S. Subashini, and N. Chidambaram. "Cardiac view classification using speed Up robust", Indian Journal of Science and Technology, Vol.8, no.7, pp.1-5,2015.
Jayashree Dev, Sanjit K Dash, Sweta Dash, and Madhusmita Swain, “A Classification Technique for Microarray Gene Expression Data using PSOFLANN‖”, International Journal on Computer Science and Engineering (IJCSE), vol.4, no.9, pp.1534-1539, 2012.
K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556 (2014).
C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going deeper with convolutions, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 1–9.
Gupta, D. J. . (2022). A Study on Various Cloud Computing Technologies, Implementation Process, Categories and Application Use in Organisation. International Journal on Future Revolution in Computer Science &Amp; Communication Engineering, 8(1), 09–12. https://doi.org/10.17762/ijfrcsce.v8i1.2064
H.-C. Shin, H. R. Roth, M. Gao, L. Lu, Z. Xu, I. Nogues, J. Yao, D. Mollura, R. M. Summers, Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning, IEEE transactions on medical imaging 35 (2016) 1285–1298.
W. Rawat, Z. Wang, Deep convolutional neural networks for image classification: A comprehensive review, Neural computation 29 (2017) 2352–2449. [
A. Serag, A. Ion-Margineanu, H. Qureshi, R. McMillan, M.-J. Saint Martin, J. Diamond, P. O’Reilly, P. Hamilton, Translational ai and deep learning in diagnostic pathology, Frontiers in Medicine 6 (2019)
N. Wahab, A. Khan, Y. S. Lee, Transfer learning based deep cnn for segmentation and detection of mitoses in breast cancer histopathological images, Microscopy 68 (2019) 216–233.
M. Yanagawa, H. Niioka, A. Hata, N. Kikuchi, O. Honda, H. Kurakami, E. Morii, M. Noguchi, Y. Watanabe, J. Miyake, et al., Application of deep learning (3-dimensional convolutional neural network) for the prediction of pathological invasiveness in lung adenocarcinoma: A preliminary study, Medicine 98 (2019).
KonstantinaKourou, Themis P. Exarchos, Konstantinos P. Exarchos, Michalis V. Karamouzis, Dimitrios I. Fotiadis, “Machine Learning Applications in Cancer Prognosis and Prediction”, ELSEVIER Computational and Structural Biotechnology Journal, vol.13, pp.8-17, 2015.
Maulik U, Chakraborty D, “Fuzzy preference based feature selection and semisupervised SVM for cancer classification”, IEEE Trans NanoBiosciences, vol.13, no.2, pp.152-160, 2014.
V. JothiPrakash and L.M.Nithya, “A Survey on Semi-Supervised Learning Techniques”, International Journal of Computer Trends and Technology (IJCTT), vol.8, no.1, pp.25-29, 2014.
Y. Celik, M. Talo, O. Yildirim, M. Karabatak, U. R. Acharya, Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images, Pattern Recognition Letters (2020).
Zhao, Yongqiang, Mohamed Reda, Kai Feng, Peng Zhang, Gaojian Cheng, ZhigangRen, Seong G. Kong, Shihan Su, HaiXia Huang, and Jiyuan Zang, “Detecting Giant Cell Cancer of Bone Lesions Using Mueller Matrix Polarization Microscopic Imaging and Multi-Parameters Fusion Network”, IEEE Sensors Journal, vol.20, no.13, pp.7208-7215, 2020.
Ho, Ngoc-Huynh, Hyung-Jeong Yang, Soo-Hyung Kim, Sung Taek Jung, and Sang-Don Joo, “Regenerative Semi-Supervised Bidirectional W-Network-Based Knee Bone Cancer Classification on Radiographs Guided by Three-Region Bone Segmentation”, IEEE Access, vol.7, pp.154277-154289, 2019.
Kiran, M. S., & Yunusova, P. (2022). Tree-Seed Programming for Modelling of Turkey Electricity Energy Demand. International Journal of Intelligent Systems and Applications in Engineering, 10(1), 142–152. https://doi.org/10.18201/ijisae.2022.278
Ellmann, Stephan, Lisa Seyler, Jochen Evers, HenrikHeinen, AlineBozec, Olaf Prante, TorstenKuwert, Michael Uder, and Tobias Baeuerle,“Prediction of early metastatic disease in experimental breast cancer bone metastasis by combining PET/CT and MRI parameters to a Model-Averaged Neural Network”, Bone, pp.254-261, 2019.
Elbashir, Murtada K, Mohamed Ezz, Mohanad Mohammed, and Said S. Saloum, “Lightweight Convolutional Neural Network for Breast Cancer Classification Using RNA-Seq Gene Expression Data”, IEEE Access, vol.7, pp.185338-185348, 2019.
Tan, Jiaxing, YongfengGao, Zhengrong Liang, Weiguo Cao, Marc J. Pomeroy, YumeiHuo, Lihong Li, Matthew A. Barish, Almas F. Abbasi, and Perry J. Pickhardt, “3D-GLCM CNN: A 3-Dimensional Gray-Level Co-Occurrence Matrix-Based CNN Model for Polyp Classification via CT Colonography”, IEEE transactions on medical imaging, vol.39, no.6, pp.2013-2024, 2019.
Dhillon, Arwinder and Ashima Singh, “eBreCaP: extreme learning-based model for breast cancer survival prediction”, IET Systems Biology, vol.14, no.3, pp.160-169, 2018.
Zhang, Baihua, Shouliang Qi, Patrice Monkam, Chen Li, Fan Yang, Yu-Dong Yao, and Wei Qian, “Ensemble learners of multiple deep CNNs for pulmonary nodules classification using CT images”, IEEE Access, vol.7, pp.110358-110371, 2018.
Linda R. Musser. (2020). Older Engineering Books are Open Educational Resources. Journal of Online Engineering Education, 11(2), 08–10. Retrieved from http://onlineengineeringeducation.com/index.php/joee/article/view/41
Kourou, Konstantina D, Vasileios C. Pezoulas, Eleni I. Georga, Themis Exarchos, Costas Papaloukas, MichalisVoulgarelis, and Andreas Goules, “Predicting Lymphoma Development by Exploiting Genetic Variants and Clinical Findings in a Machine Learning-Based Methodology With Ensemble Classifiers in a Cohort of Sjögren's Syndrome Patients”, IEEE Open Journal of Engineering in Medicine and Biology, vol.1, pp.49-56, 2018.
Yin, Tang-Kai and Nan-TsingChiu, “A computer-aided diagnosis for locating abnormalities in bone scintigraphy by a fuzzy system with a three-step minimization approach”, IEEE Transactions on Medical Imaging, vol.23, no.5, pp.639-654, 2014.
Anand, D., Arulselvi, G., Balaji, G.N. (2022). An Assessment on Bone Cancer Detection Using Various Techniques in Image Processing. In: Deepak, B.B.V.L., Parhi, D., Biswal, B., Jena, P.C. (eds) Applications of Computational Methods in Manufacturing and Product Design. Lecture Notes in Mechanical Engineering.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





