Wrapper Fuzzy Approach with 3d Fast Convolution Neural Network (FCNN) Based Feature Selection in Protein Sequence Classification
Keywords:
Bioinformatics, protein sequences, classification, feature selection, noise removal, wrapper fuzzy, Classification using 3D- Fast Convolution neural network (3D- FCNN)Abstract
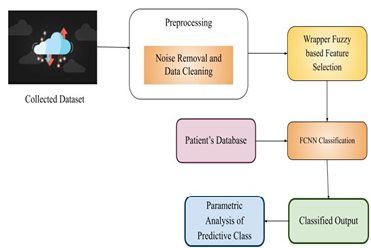
In research area, an emerging field is Bioinformatics in the past decades. Biological data storage and management was the definite motivation of bioinformatics and the tools for computation are developed and analyzed for enhancing their understanding. The data size is gathered under different project sequence is exponentially increased, that provides the problems for the methods of experiment. Newly sequenced protein and known functions proteins have gap and this gap is reduced by several techniques of computation incorporating classification and algorithms of clustering were presented in the past. The sequences of protein are classified into superfamilies exists in literature is useful for the prediction of structure and function of huge proteins that are discovered newly. The existing classification’s results are unacceptable because of larger feature size acquired by several approaches of feature encoding. This paper proposes noise removal technique depending on selection of feature for protein sequence classification. Here we use wrapper fuzzy model with fast convolution neural network (FCNN) for feature selection and remove the noise. This research involved in removal of noisy or unwanted data related to protein composite. To improve classification accuracy, wrapper fuzzy is utilized for selection of features. Wrapper algorithm involved in selection of protein features for accurate identification of protein composites. For classification we use 3D FCNN which can improve the accuracy of classification. The classification of protein proposed in this method proves momentous enhancement with respect to measuring the metrics of performance: accuracy, sensitivity, specificity, recall, F-measure, and etc.
Downloads
References
Xing, Zhengzheng, Jian Pei, and Eamonn Keogh. "A brief survey on sequence classification." ACM Sigkdd Explorations Newsletter 12.1 (2010): 40-48.
Cai, Jie, et al. "Feature selection in machine learning: A new perspective." Neurocomputing 300 (2018): 70-79.
Wang, Lipo, Yaoli Wang, and Qing Chang. "Feature selection methods for big data bioinformatics: A survey from the search perspective." Methods 111 (2016): 21-31.
Yang, Wuritu, et al. "A brief survey of machine learning methods in protein sub-Golgi localization." Current Bioinformatics 14.3 (2019): 234-240.
Saeys, Yvan, InakiInza, and Pedro Larranaga. "A review of feature selection techniques in bioinformatics." bioinformatics 23.19 (2007): 2507-2517.
Nemati, Shahla, et al. "A novel ACO–GA hybrid algorithm for feature selection in protein function prediction." Expert systems with applications 36.10 (2009): 12086-12094.
Ma, Shuangge, and Jian Huang. "Penalized feature selection and classification in bioinformatics." Briefings in bioinformatics 9.5 (2008): 392-403.
Qin, Xinyi, et al. "Structural protein fold recognition based on secondary structure and evolutionary information using machine learning algorithms." Computational Biology and Chemistry 91 (2021): 107456.
Zhao, Xudong, et al. "ECFS-DEA: an ensemble classifier-based feature selection for differential expression analysis on expression profiles." BMC bioinformatics 21.1 (2020): 43.
Guannoni, Naoual, FaouziMhamdi, and MouradElloumi. "Improved Feature Selection Algorithm for Biological Sequences Classification." International Conference on Knowledge Science, Engineering and Management. Springer, Cham, 2019.
Sang, Xiuzhi, et al. "HMMPred: accurate Prediction of DNA-binding proteins based on HMM Profiles and XGBoost feature selection." Computational and mathematical methods in medicine 2020 (2020).
Sarkar, Jnanendra Prasad, et al. "Machine learning integrated ensemble of feature selection methods followed by survival analysis for predicting breast cancer subtype specific miRNA biomarkers." Computers in Biology and Medicine 131 (2021): 104244.
Cinelli, Mattia, et al. "Feature selection using a one dimensional naïve Bayes’ classifier increases the accuracy of support vector machine classification of CDR3 repertoires." Bioinformatics 33.7 (2017): 951-955.
Blanco, Jose Liñares, et al. "Prediction of high anti-angiogenic activity peptides in silico using a generalized linear model and feature selection." Scientific reports 8.1 (2018): 1-11.
Rangasamy, Ranjani Rani, and RamyachitraDuraisamy. "Ensemble of Artificial Bee Colony Optimization and Random Forest Technique for Feature Selection and Classification of Protein Function Family Prediction." Soft Computing in Data Analytics. Springer, Singapore, 2019. 165-173.
Sequeira, Ana Marta, Diana Lousa, and Miguel Rocha. "ProPythia: A Python Automated Platform for the Classification of Proteins Using Machine Learning." International Conference on Practical Applications of Computational Biology & Bioinformatics. Springer, Cham, 2020.
Saikumar, K., Rajesh, V., Babu, B.S. (2022). Heart disease detection based on feature fusion technique with augmented classification using deep learning technology. Traitement du Signal, Vol. 39, No. 1, pp. 31-42. https://doi.org/10.18280/ts.390104.
Kailasam, S., Achanta, S.D.M., Rama Koteswara Rao, P., Vatambeti, R., Kayam, S. (2022). An IoT-based agriculture maintenance using pervasive computing with machine learning technique. International Journal of Intelligent Computing and Cybernetics, 15(2), pp. 184–197.
Rao, K. S., Reddy, B. V., Sarada, K., & Saikumar, K. (2021). A Sequential Data Mining Technique for Identification of Fault Zone Using FACTS-Based Transmission. In Handbook of Research on Innovations and Applications of AI, IoT, and Cognitive Technologies (pp. 408-419). IGI Global.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 T. Sudha Rani, A. Yesu Babu, D. Haritha

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





