Design Development of Machine Learning Secure Image Transmission Based Cooperative Communication and Gaussian Elimination
Keywords:
Cooperative Communication, MIMO, MRI Images, Machine Learning, Gaussian Elimination, K Means, KNNAbstract
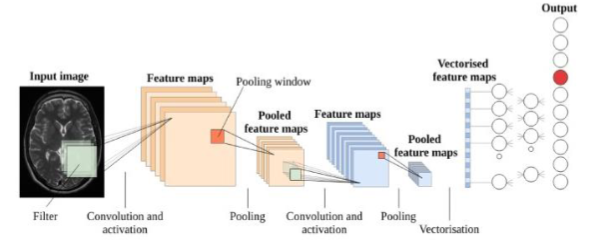
Tumors are difficult to notice in medical imaging due to their intricate structure and noise, making it difficult and time-consuming for doctors to locate them. This is important since locating and pinpointing the tumor’s site at an early stage is critical. Due to the complicated structure of tumors and the involution of noise in Magnetic Resonance Imaging (MRI) data, physical tumors identification has become a difficult and time-consuming process for medical practitioners. Thus, this paper proposes a machine learning-based approach for segmenting and categorizing of magnetic resonance images to identify brain tumors. The framework of this paper uses SVM and Naive Bayes algorithms for image pre-processing, feature extraction, and classification. The results indicated that the two classification algorithms used (Naïve Bayes and SVM) had an accuracy of 0.89 for SVM and 0.51 for naïve Bayes, a sensitivity of 0.57 and 0.85, and a specificity of 0.99 0.42, respectively. The findings indicate that SVM is more precise and specified than Naïve Bayes but that Naïve Bayes is more sensitive, with a sensitivity of 0.85. The Naïve Bayes classifier produces modest performance when compared to SVM classifiers.
Downloads
References
S. Bauer, R. Wiest, L. P. Nolte, and M. Reyes, “A survey of MRI-based medical image analysis for brain tumor studies,” Phys. Med. Biol., vol. 58, no. 13, Jul. 2013, doi: 10.1088/0031-9155/58/13/R97.
G. Mohan and M. M. Subashini, “MRI based medical image analysis: Survey on brain tumor grade classification,” Biomed. Signal Process. Control, vol. 39, pp. 139–161, Jan. 2018, doi: 10.1016/J.BSPC.2017.07.007.
B. H. Menze et al., “The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS),” IEEE Trans. Med. Imaging, vol. 34, no. 10, pp. 1993–2024, Oct. 2015, doi: 10.1109/TMI.2014.2377694.
J. Cheng et al., “Enhanced Performance of Brain Tumor Classification via Tumor Region Augmentation and Partition,” PLoS One, vol. 10, no. 10, p. e0140381, Oct. 2015, doi: 10.1371/JOURNAL.PONE.0140381.
G. Litjens et al., “A survey on deep learning in medical image analysis,” Med. Image Anal., vol. 42, pp. 60–88, Dec. 2017, doi: 10.1016/J.MEDIA.2017.07.005.
J. Sachdeva, V. Kumar, I. Gupta, N. Khandelwal, and C. K. Ahuja, “A package-SFERCB- ‘Segmentation, feature extraction, reduction and classification analysis by both SVM and ANN for brain tumors,’” Appl. Soft Comput., vol. 47, pp. 151–167, Oct. 2016, doi: 10.1016/J.ASOC.2016.05.020.
S.-L. Jui et al., ‘Brain MRI Tumor Segmentation with 3D Intracranial Structure Deformation Features’, Intelligent Systems, IEEE, vol. 31, pp. 66–76, Mar. 2016.
A. Makropoulos et al., “Automatic whole brain MRI segmentation of the developing neonatal brain,” IEEE Trans. Med. Imaging, vol. 33, no. 9, pp. 1818–1831, 2014, doi: 10.1109/TMI.2014.2322280.
M. Ravikumar, B. J. Shivaprasad, and D. S. Guru, “Enhancement of MRI Brain Images Using Fuzzy Logic Approach,” Commun. Comput. Inf. Sci., vol. 1381 CCIS, pp. 131–137, 2021, doi: 10.1007/978-981-16-0493-5_12.
Z. Iqbal, M. A. Khan, M. Sharif, J. H. Shah, M. H. ur Rehman, and K. Javed, “An automated detection and classification of citrus plant diseases using image processing techniques: A review,” Comput. Electron. Agric., vol. 153, pp. 12–32, Oct. 2018, doi: 10.1016/J.COMPAG.2018.07.032.
E. S. Biratu, F. Schwenker, Y. M. Ayano, and T. G. Debelee, “A Survey of Brain Tumor Segmentation and Classification Algorithms,” J. imaging, vol. 7, no. 9, Sep. 2021, doi: 10.3390/JIMAGING7090179.
R. V. Suryavamsi, L. S. T. Reddy, S. Saladi, and Y. Karuna, “Comparative Analysis of Various Enhancement Methods for Astrocytoma MRI Images,” Proc. 2018 IEEE Int. Conf. Commun. Signal Process. ICCSP 2018, pp. 812–816, Nov. 2018, doi: 10.1109/ICCSP.2018.8524441.
B. De Blasi et al., “Pipeline comparison for the pre-processing of resting-state data in epilepsy,” Eur. Signal Process. Conf., vol. 2018-September, pp. 1137–1141, Nov. 2018, doi: 10.23919/EUSIPCO.2018.8553119.
W. Zhao and J. Wang, “A new method of the forest dynamic inspection color image sharpening process,” ICACTE 2010 - 2010 3rd Int. Conf. Adv. Comput. Theory Eng. Proc., vol. 4, 2010, doi: 10.1109/ICACTE.2010.5579715.
S. Widyarto, S. R. B. Kassim, and W. K. Sari, “2D-sigmoid enhancement prior to segment MRI glioma tumour: Pre-image-processing,” Int. Conf. Electr. Eng. Comput. Sci. Informatics, vol. 2017-December, Dec. 2017, doi: 10.1109/EECSI.2017.8239103.
S. Poornachandra and C. Naveena, “Pre-processing of MR Images for Efficient Quantitative Image Analysis Using Deep Learning Techniques,” Proc. - 2017 Int. Conf. Recent Adv. Electron. Commun. Technol. ICRAECT 2017, pp. 191–195, Oct. 2017, doi: 10.1109/ICRAECT.2017.43.
J. Zhang, M. Liu, L. An, Y. Gao, and D. Shen, “Alzheimer’s disease diagnosis using landmark-based features from longitudinal structural MR images,” IEEE J. Biomed. Heal. Informatics, vol. 21, no. 6, pp. 1607–1616, Nov. 2017, doi: 10.1109/JBHI.2017.2704614.
F. H. Almukhtar, “Principal Component Analysis and K-Nearest Neighbor Classifier for Facial Expression Recognition System,” Int. J. Curr. Res. Acad. Rev., vol. 6, no. 5, pp. 17–20, May 2018, doi: 10.20546/IJCRAR.2018.605.004.
M. Osadebey, M. Pedersen, D. Arnold, and K. Wendel-Mitoraj, ‘No-Reference Quality Measure in Brain MRI Images using Binary Operations, Texture and Set Analysis’, IET Image Processing, vol. 11, pp. 672–684, Sep. 2017, doi: 10.1049/iet-ipr.2016.0560.
H.-Y. Tsai, H. Zhang, C.-L. Hung, and G. Min, ‘GPU-Accelerated Features Extraction From Magnetic Resonance Images’, IEEE Access, vol. 5, pp. 22634–22646, 2017, doi: 10.1109/ACCESS.2017.2756624.
S. Pereira, A. Pinto, V. Alves, and C. A. Silva, “Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images,” IEEE Trans. Med. Imaging, vol. 35, no. 5, pp. 1240–1251, May 2016, doi: 10.1109/TMI.2016.2538465.
J. Kim, C. Lenglet, Y. Duchin, G. Sapiro, and N. Harel, “SEMI-AUTOMATIC SEGMENTATION OF BRAIN SUBCORTICAL STRUCTURES FROM HIGH-FIELD MRI,” IEEE J. Biomed. Heal. informatics, vol. 18, no. 5, p. 1678, Sep. 2014, doi: 10.1109/JBHI.2013.2292858.
M. Mittal, M. Arora, T. Pandey, and L. M. Goyal, “Image Segmentation Using Deep Learning Techniques in Medical Images,” pp. 41–63, 2020, doi: 10.1007/978-981-15-1100-4_3.
B. N. Mohammed, F. H. Al-Mukhtar, R. Z. Yousif, and Y. S. Almashhadani, “Automatic Classification of Covid-19 Chest X-Ray Images Using Local Binary Pattern and Binary Particle Swarm Optimization for Feature Selection,” Cihan Univ. Sci. J., vol. 5, no. 2, pp. 46–51, Nov. 2021, doi: 10.24086/CUESJ.V5N2Y2021.PP46-51.
Z. Deng, X. Zhu, D. Cheng, M. Zong, and S. Zhang, “Efficient kNN classification algorithm for big data,” Neurocomputing, vol. 195, pp. 143–148, Jun. 2016, doi: 10.1016/J.NEUCOM.2015.08.112.
E. Suryawati et al., “Unsupervised feature learning-based encoder and adversarial networks,” J. Big Data, vol. 8, no. 1, pp. 1–17, Dec. 2021, doi: 10.1186/S40537-021-00508-9/TABLES/4.
M. Galety, F. H. Al Mukthar, R. J. Maaroof, and F. Rofoo, “Deep Neural Network Concepts for Classification using Convolutional Neural Network: A Systematic Review and Evaluation,” Tech. Rom. J. Appl. Sci. Technol., vol. 3, no. 8, pp. 58–70, Sep. 2021, doi: 10.47577/TECHNIUM.V3I8.4554.
Z. H. Moe, T. San, M. M. Khin, and H. M. Tin, “Comparison of Naive Bayes and Support Vector Machine Classifiers on Document Classification,” 2018 IEEE 7th Glob. Conf. Consum. Electron. GCCE 2018, pp. 285–286, Dec. 2018, doi: 10.1109/GCCE.2018.8574785.
S. H. Ismael, S. W. Kareem, and F. H. Almukhtar, “Medical Image Classification Using Different Machine Learning Algorithms,” Mosul Univ., vol. 14, no. 1, pp. 135–147, May 2020, doi: 10.33899/CSMJ.2020.164682.
Z. Lai and H. Deng, “Medical Image Classification Based on Deep Features Extracted by Deep Model and Statistic Feature Fusion with Multilayer Perceptron ,” Comput. Intell. Neurosci., vol. 2018, 2018, doi: 10.1155/2018/2061516.
N. Xu, W. Shan, J. Qi, J. Wu, and Q. Wang, “Presurgical Evaluation of Epilepsy Using Resting-State MEG Functional Connectivity,” Front. Hum. Neurosci., vol. 15, p. 364, Jul. 2021, doi: 10.3389/FNHUM.2021.649074/BIBTEX.
D. Y. Mikhail, F. H. Al-Mukhtar, and S. W. Kareem, “A Comparative Evaluation of Cancer Classification via TP53 Gene Mutations Using Machin Learning,” Asian Pacific J. Cancer Prev., vol. 23, no. 7, pp. 2459–2467, Jul. 2022, doi: 10.31557/APJCP.2022.23.7.2459.
M. Z. AL-Dabagh and D. F. H. AL-Mukhtar, “Breast Cancer Diagnostic System Based on MR images Using KPCA-Wavelet Transform and Support Vector Machine,” Int. J. Adv. Eng. Res. Sci., vol. 4, no. 3, pp. 258–263, 2017, doi: 10.22161/IJAERS.4.3.41.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Firas Husham Almukhtar

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





