3 Phase Atrous Net with DCO-3DSPMRINET Model for Scoliosis Prediction
Keywords:
Intervertebral disc, scoliotic, quadruple up sampling operation, 3 phase atrous net, 3DSpMRINet, accuracy, omnidirectional sagittal block matching algorithm.Abstract
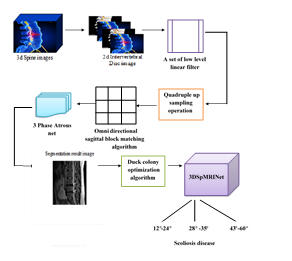
Intervertebral Disc (ID) is the mattress like structure that holds the bones of the spine together thus these discs increase the stability of the spinal column and ID images are used in the prediction of Scoliosis disease. However, while processing these images existing techniques use edge operators to locate four points on vertebral body for prediction of disc bulge but it is very difficult to obtain such image because the severity in the selected plane image is still uncertain. Hence a novel Quadruple Up Sampling Operation up sample the images double times with the Omnidirectional sagittal block matching algorithm that select, match and label the image hence, the intervertebral disc's severity and the plane image as a slice of the present frame remain predictable. Moreover, during segmentation it is impossible for 3D structure to reconstruct automatically due to over segmentation and it provide only less detail about disease growing rate. Hence the technique 3 phase Atrous Net automatically segment and predict the sub image pair by segmentation and elastic atlas mapping with the cascade and parallel atrous convolution thus provide separation between vertebral disc near together and overcome over segmentation with the Duck colony optimization algorithm for feature selection. However, in classification stage sematic and discreet reconstruction limit the detail in image feature extraction so it results in perplexing outcome. Hence 3DSpMRINet technique uses sparse learning that provide feature weight vector that enhances the quality of the image and provide a high accuracy of disease classification. The proposed model for scoliosis prediction has been implemented in python platform and the result shows improved accuracy, recall rate, precision and F1 score.
Downloads
References
R. Kritschil, M. Scott, G. Sowa, and N. Vo, “Role of autophagy in intervertebral disc degeneration,” Journal of cellular physiology, vol. 237, no. 2, pp.1266-1284, 2022.
D. Sakai, J. Schol and M. Watanabe, “Clinical Development of Regenerative Medicine Targeted for Intervertebral Disc Disease,” Medicina, vol. 58, no. 2, pp. 267, 2022.
G. Bjornsdottir, L. Stefansdottir, G. Thorleifsson, P. Sulem, K. Norland, E. Ferkingstad, A. Oddsson, F. Zink, S.H. Lund, M.S. Nawaz and G. Bragi Walters, “Rare SLC13A1 variants associate with intervertebral disc disorder highlighting role of sulfate in disc pathology,” Nature communications, vol. 13, no. 1, pp.1-13, 2022.
T.J. DiStefano, K. Vaso, G. Danias, H.N. Chionuma, J.R. Weiser and J.C. Iatridis, “Extracellular vesicles as an emerging treatment option for intervertebral disc degeneration: therapeutic potential, translational pathways, and regulatory considerations,” Advanced Healthcare Materials, vol. 11, no. 5, pp. 2100596, 2022.
J. Zieba, K.N. Forlenza, K. Heard, J.H. Martin, M. Bosakova, D.H. Cohn, S.P. Robertson, P. Krejci, and D. Krakow, “Intervertebral disc degeneration is rescued by TGFβ/BMP signaling modulation in an ex vivo filamin B mouse model,” Bone Research, vol. 10, no. 1, pp.1-12, 2022.
S. Chen, L. Lei, Z. Li, F. Chen, Y. Huang, G. Jiang, X. Guo, Z. Zhao, H. Liu, H. Wang and C. Liu, “Grem1 accelerates nucleus pulposus cell apoptosis and intervertebral disc degeneration by inhibiting TGF-β-mediated Smad2/3 phosphorylation,” Experimental & Molecular Medicine, pp.1-13, 2022.
M. Yang, D. Xiang, Y. Chen, Y. Cui, S. Wang, and W. Liu, “An Artificial PVA-BC Composite That Mimics the Biomechanical Properties and Structure of a Natural Intervertebral Disc,” Materials, vol. 15, no. 4, pp.1481, 2022.
Z. Wang, H. Chen, Q. Tan, J. Huang, S. Zhou, F. Luo, D. Zhang, J. Yang, C. Li, B. Chen, and X. Sun, “Inhibition of aberrant Hif1α activation delays intervertebral disc degeneration in adult mice,” Bone Research, vol. 10, no. 1, pp.1-16, 2022.
C. Wang, S. Guo, Q. Gu, X. Wang, L. Long, C. Xiao, M. Xie, H. Shen and S. Li, “Exosomes: A promising therapeutic strategy for intervertebral disc degeneration,” Experimental Gerontology, pp.111806, 2022.
T. Ohnishi, N. Iwasaki, and H. Sudo, “Causes of and Molecular Targets for the Treatment of Intervertebral Disc Degeneration: A Review,” Cells, vol. 11, no. 3, pp. 394, 2022.
X. Bai, M. Jiang, J. Wang, S. Yang, Z. Liu, H. Zhang, and X. Zhu, “Cyanidin attenuates the apoptosis of rat nucleus pulposus cells and the degeneration of intervertebral disc via the JAK2/STAT3 signal pathway in vitro and in vivo,” Pharmaceutical Biology, vol. 60, no. 1, pp.427-436, 2022.
Y. Fan, L. Zhao, Y. Lai, K. Lu, and J. Huang, “CRISPR/Cas9-Mediated Loss-of-Function of β-Catenin Attenuates Intervertebral Disc Degeneration,” Molecular Therapy-Nucleic Acids, 2022.
V. Francisco, J. Pino, M.Á. González-Gay, F. Lago, J. Karppinen, O. Tervonen, A. Mobasheri, and O. Gualillo, “A new immunometabolic perspective of intervertebral disc degeneration,” Nature Reviews Rheumatology, vol. 18, no. 1, pp.47-60, 2022.
T. Wu, X. Jia, Z. Zhu, K. Guo, Q. Wang, Z. Gao, X. Li, Y. Huang, and D. Wu, “Inhibition of miR-130b-3p restores autophagy and attenuates intervertebral disc degeneration through mediating ATG14 and PRKAA1,” Apoptosis, pp.1-17, 2022.
F. Mahyudin, C.R.S. Prakoeswa, H.B. Notobroto, D. Tinduh, R. Ausrin, F.A. Rantam, H. Suroto, D.N. Utomo, and S. Rhatomy, “An update of current therapeutic approach for Intervertebral Disc Degeneration: A review article,” Annals of Medicine and Surgery, pp.103619, 2022.
K. Miyazaki, S. Miyazaki, T. Yurube, Y. Takeoka, Y. Kanda, Z. Zhang, Y. Kakiuchi, R. Tsujimoto, H. Ohnishi, T. Matsuo, and M. Ryu, “Protective Effects of Growth Differentiation Factor-6 on the Intervertebral Disc: An In Vitro and In Vivo Study,” Cells, vol. 11, no. 7, pp.1174, 2022.
K. Yamada, N. Iwasaki, and H. Sudo, “Biomaterials and Cell-Based Regenerative Therapies for Intervertebral Disc Degeneration with a Focus on Biological and Biomechanical Functional Repair: Targeting Treatments for Disc Herniation,” Cells, vol. 11, no. 4, pp. 602, 2022.
M.P. Culbert, J.P. Warren, A.R. Dixon, H.L. Fermor, P.A. Beales, and R.K. Wilcox, “Evaluation of injectable nucleus augmentation materials for the treatment of intervertebral disc degeneration,” Biomaterials Science, 2022.
S. Khalid, S. Ekram, A. Salim, G.R. Chaudhry, and I. Khan, “Transcription regulators differentiate mesenchymal stem cells into chondroprogenitors, and their in vivo implantation regenerated the intervertebral disc degeneration,” World Journal of Stem Cells, vol. 14, no. 2, pp.163, 2022.
H. Liang, R. Luo, G. Li, W. Zhang, Y. Song, and C. Yang, “The Proteolysis of ECM in Intervertebral Disc Degeneration,” International Journal of Molecular Sciences, vol. 23, no. 3, pp.1715, 2022.
C. Wang, L. Cui, Q. Gu, S. Guo, B. Zhu, X. Liu, Y. Li, X. Liu, D. Wang, and S. Li, “The Mechanism and Function of miRNA in Intervertebral Disc Degeneration,” Orthopaedic Surgery, vol. 14, no. 3, pp.463-471, 2022.
Y. Wang, J. Kang, X. Guo, D. Zhu, M. Liu, L. Yang, G. Zhang, and X. Kang, “Intervertebral disc degeneration models for pathophysiology and regenerative therapy-benefits and limitations,” Journal of Investigative Surgery, vol. 35, no. 4, pp.935-952, 2022.
S. Ashwinkumar, S. Rajagopal, V. Manimaran, and B. Jegajothi, “Automated plant leaf disease detection and classification using optimal MobileNet based convolutional neural networks,” Materials Today: Proceedings, vol. 51, pp.480-487, 2022.
S. Shastri, I. Kansal, S. Kumar, K. Singh, R. Popli, and V. Mansotra, “CheXImageNet: a novel architecture for accurate classification of Covid-19 with chest x-ray digital images using deep convolutional neural networks,” Health and Technology, pp.1-12, 2022.
F. Rehman, S.I.A. Shah, N. Riaz, and S.O. Gilani, “A robust scheme of vertebrae segmentation for medical diagnosis,” IEEE Access, vol. 7, pp.120387-120398, 2019.
E.J. Hwang, S. Kim, and J.Y. Jung, “Bone Marrow Radiomics of T1-Weighted Lumber Spinal MRI to Identify Diffuse Hematologic Marrow Diseases: Comparison With Human Readings,” IEEE Access, vol. 8, pp.133321-133329, 2020.
F. Fallah, S.S. Walter, F. Bamberg, and B. Yang, “Simultaneous volumetric segmentation of vertebral bodies and intervertebral discs on fat-water MR images,” IEEE journal of biomedical and health informatics, vol. 23, no. 4, pp.1692-1701, 2018.
L. Lin, X. Tao, W. Yang, S. Pang, Z. Su, H. Lu, S. Li, Q. Feng, and B. Chen, “Quantifying Axial Spine Images Using Object-Specific Bi-Path Network,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 8, pp.2978-2987, 2021.
S. Pang, C. Pang, L. Zhao, Y. Chen, Z. Su, Y. Zhou, M. Huang, W. Yang, H. Lu, and Q. Feng, “Spineparsenet: spine parsing for volumetric MR image by a two-stage segmentation framework with semantic image representation,” IEEE Transactions on Medical Imaging, vol. 40, no. 1, pp.262-273, 2020.
X. Li, Q. Dou, H. Chen, C.W. Fu, X. Qi, D.L. Belavý, G. Armbrecht, D. Felsenberg, G. Zheng, and P.A. Heng, “3D multi-scale FCN with random modality voxel dropout learning for intervertebral disc localization and segmentation from multi-modality MR images,” Medical image analysis, vol. 45, pp.41-54, 2018.
H. Wang, T. Zhang, K.M.C. Cheung, and G.K.H. Shea, “Application of deep learning upon spinal radiographs to predict progression in adolescent idiopathic scoliosis at first clinic visit,” EClinicalMedicine, vol. 42, pp.101220, 2021.
M.L. Nault, M. Beauséjour, M. Roy-Beaudry, J.M. Mac-Thiong, J. de Guise, H. Labelle, and S. Parent, “A predictive model of progression for adolescent idiopathic scoliosis based on 3D spine parameters at first visit,” Spine, vol. 45, no. 9, pp.605-611, 2020.
J. Zhang, K.Y. Cheuk, L. Xu, Y. Wang, Z. Feng, T. Sit, K.L. Cheng, E. Nepotchatykh, T.P. Lam, Z. Liu, and A.L. Hung, “A validated composite model to predict risk of curve progression in adolescent idiopathic scoliosis,” EClinicalMedicine, vol. 18, p.100236, 2020.
J. Yi, P. Wu, Q. Huang, H. Qu, and D.N. Metaxas, “Vertebra-focused landmark detection for scoliosis assessment,” In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). IEEE, pp. 736-740, 2020, April.
J. Bradley, and S. Rajendran, “Developing predictive models for early detection of intervertebral disc degeneration risk,” Healthcare Analytics, vol. 2, pp. 100054, 2022.
S. Ma, Y. Huang, X. Che, and R. Gu, “Faster RCNN‐based detection of cervical spinal cord injury and disc degeneration,” Journal of Applied Clinical Medical Physics, vol. 21, no. 9, pp.235-243, 2020.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





