Fusing Expert Knowledge and Deep Learning for Accurate Cervical Cancer Diagnosis in Pap Smear Images: A Multiscale U-Net with Fuzzy Automata
Keywords:
Segmentation, Fuzzy Automata, Pap smear images, Multiscale U-NetAbstract
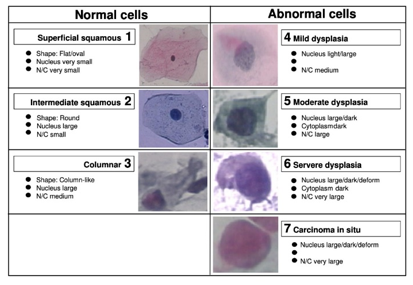
Ovarian cancer is a severe disease that impacts many women in developing countries. Increasing screening capacity is the most effective strategy for lowering cancer risk and saving people's lives. Early stages of cervical cancer often lack symptoms, making it the fourth leading cause of mortality among women. Although cancer cells grow slowly in the cervix and can be effectively treated if detected early, detecting it before it rapidly spreads are a major challenge for the medical community. Segmentation is a critical screening step as it enhances our comprehension of cell morphological properties. This study provides a technique to segment multi-class cells into Nucleus and Cytoplasmic areas. Multi-resolution U-Net (MRU-Net) is provided for medical image segmentation to bypass the constraints of U-convolution Net's kernel with a restricted receptive field and undetermined ideal network width. First, additional semantic information is extracted from the images using a series of recurrent convolutions. Second, to distinguish the characteristics, a convolutional unit with distinct receptive fields is utilized. The effects of network width inconsistency may be mitigated by integrating a convolution layer with a large number of receptive fields. The effectiveness of the research was measured against state-of-the-art methods using the Herlev dataset and classification structures were used to get excellent results. Effectiveness indicators for both groups suggest that the method is reliable enough to complete the task. The approach may enable doctors to identify cervical cell anomalies and provide improved medical care. MRU-Net is evaluated using varied medical image segmentation datasets.
Downloads
References
F. J., E. M., L. F., C. M., Global cancer observatory: Cancer today, 2020, URL https://gco.iarc.fr/today/.
H. Sung, J. Ferlay, R.L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: Cancer J. Clin. 71 (3) (2021) http://dx.doi.org/10.3322/caac.21660.
R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 2019, CA: Cancer J. Clin. 69 (1) (2019) 7–34, http://dx.doi.org/10.3322/caac.21551.
M. Safaeian, D. Solomon, P.E. Castle, Cervical cancer prevention—Cervical screening: Science in evolution, Obstet. Gynecol. Clin. North Am. 34 (4) (2007) 739–760, http://dx.doi.org/10.1016/j.ogc.2007.09.004.
P. Bamford, B. Lovell, Unsupervised cell nucleus segmentation with active contours, Signal Process. 71 (2) (1998) 203–213, http://dx.doi.org/10.1016/ S0165-1684(98)00145-5.
L. Zhang, H. Kong, C.T. Chin, S. Liu, Z. Chen, T. Wang, S. Chen, Segmentation of cytoplasm and nuclei of abnormal cells in cervical cytology using global and local graph cuts, Comput. Med. Imaging Graph. (2014) http://dx.doi.org/10.1016/j. compmedimag.2014.02.001.
K.P. Win, Y. Kitjaidure, K. Hamamoto, T.M. Aung, Computer-assisted screening for cervical cancer using digital image processing of pap smear images, Appl. Sci. (Switz.) 10 (5) (2020) http://dx.doi.org/10.3390/app10051800.
M.E. Plissiti, C. Nikou, A. Charchanti, Automated detection of cell nuclei in Pap smear images using morphological reconstruction and clustering, IEEE Trans. Inf. Technol. Biomed. 15 (2) (2011) 233–241, http://dx.doi.org/10.1109/TITB.2010. 2087030.
B. Sharma, K.K. Mangat, An improved nucleus segmentation for cervical cell images using FCM clustering and BPNN, in: 2016 International Conference on Advances in Computing, Communications and Informatics, ICACCI 2016, 2016, pp. 1924–1929, http://dx.doi.org/10.1109/ICACCI.2016.7732332.
T. Chankong, N. Theera-Umpon, S. Auephanwiriyakul, Automatic cervical cell segmentation and classification in Pap smears, Comput. Methods Programs Biomed. 113 (2) (2014) 539–556, http://dx.doi.org/10.1016/j.cmpb.2013.12. 012.
K. Li, Z. Lu, W. Liu, J. Yin, Cytoplasm and nucleus segmentation in cervical smear images using Radiating GVF Snake, Pattern Recognit. 45 (4) (2012) 1255–1264, http://dx.doi.org/10.1016/j.patcog.2011.09.018.
H. Bandyopadhyay, M. Nasipuri, Segmentation of pap smear images for cervical cancer detection, in: 2020 IEEE Calcutta Conference, CALCON 2020 - Proceedings, 2020, pp. 30–33, http://dx.doi.org/10.1109/CALCON49167.2020. 9106484.
W. Wasswa, J. Obungoloch, A.H. Basaza-Ejiri, A. Ware, Automated segmentation of nucleus, cytoplasm and background of cervical cells from pap-smear images using a trainable pixel level classifier, in: Proceedings - Applied Imagery Pattern Recognition Workshop 2019-Octob, I, 2019, http://dx.doi.org/10.1109/ AIPR47015.2019.9174599.
A.M. Braga, R.C. Marques, F.N. Medeiros, J.F.R. Neto, A.G. Bianchi, C.M. Carneiro, D.M. Ushizima, Hierarchical median narrow band for level set segmentation of cervical cell nuclei, Measurement 176 (2021) 109232, http://dx. doi.org/10.1016/j.measurement.2021.109232.
Y. Song, L. Zhang, S. Chen, D. Ni, B. Li, Y. Zhou, B. Lei, T. Wang, A deep learning based framework for accurate segmentation of cervical cytoplasm and nuclei, in: Conference Proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference 2014, 2014, pp. 2903–2906, http: //dx.doi.org/10.1109/EMBC.2014.6944230.
J. Zhao, Q. Li, X. Li, H. Li, L. Zhangs, Automated segmentation of cervical nuclei in pap smear images using deformable multi-path ensemble model, in: Proceedings - International Symposium on Biomedical Imaging 2019, 2019, pp. 1514–1518, http://dx.doi.org/10.1109/ISBI.2019.8759262, arXiv:1812.00527.
S. Gautam, H.K. K, A. Jith, A. Bhavsar, N. Natarajan, Considerations for a PAP smear image analysis system with CNN features, 2018, pp. 1–8, arXiv: 1806.09025. URL http://arxiv.org/abs/1806.09025.
Kurnianingsih, K.H.S. Allehaibi, L.E. Nugroho, Widyawan, L. Lazuardi, A.S. Prabuwono, T. Mantoro, Segmentation and classification of cervical cells using deep learning, IEEE Access 7 (2019) 116925–116941, http://dx.doi.org/10.1109/ ACCESS.2019.2936017.
A. Tareef, Y. Song, W. Cai, D.D. Feng, M. Chen, Automated three-stage nucleus and cytoplasm segmentation of overlapping cells, in: 2014 13th International Conference on Control Automation Robotics and Vision, ICARCV 2014 2014 (December), 2014, pp. 865–870, http://dx.doi.org/10.1109/ICARCV.2014. 7064418.
J. Jantzen, J. Norup, G. Dounias, B. Bjerregaard, Pap-smear benchmark data for pattern classification, 2005, pp. 1–9, URL https://orbit.dtu.dk/en/publications/ pap-smear-benchmark-data-for-pattern-classification.
G. Dounias, MDE-Lab: The Management and Decision Engineering Laboratory. URL http://mde-lab.aegean.gr/.
Zhang, Deyu, Liu, Jinhuai MSU-Net: Multi-Scale U-Net for 2D Medical Image Segmentation, Frontiers in Genetics https://www.frontiersin.org/articles/10.3389/fgene.2021.639930
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





