Identifying Biomarkers from Medical Images Using Machine Learning Techniques
Keywords:
convolutional neural networks, biomarker identification, auto encodersAbstract
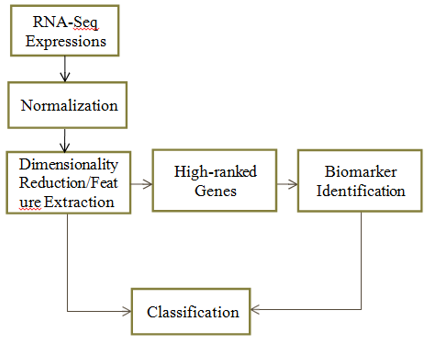
Genetic information is necessary for studying the biological processes that, when interrupted, cause certain cancers to form. Although advances in sequencing technology have made it possible to record the nuances of gene interaction in several data formats, using these methods to identify, diagnose, and treat cancer remains difficult. Machine learning has helped researchers in a number of areas, including supervised and unsupervised learning, as well as gene identification, but the results have been less than visually satisfying. Using RNA-SEQ data from The Cancer Genome Atlas, this research focuses on multi-class classification of cancer, extraction of key characteristics, and identification of relevant genes for 10 different types of cancer.Tests conducted with the restricted hardware resources at hand have shown that these limitations do not always exclude the possibility of positive results. Stacked de-noising auto encoders were employed for feature extraction and biomarker identification, while 1D convolutional neural networks were used for classification. Both the recovered features and the relevant genes were used in the classification process, with the former typically performing better (about 94% accurate) than the latter (95% accurate). By using stacked denoising auto-encoders to construct matrix weights and features, we were able to identify common cancer-related pathways and their associated genes.
Downloads
References
Gupta A, Wang H, Ganapathiraju M. Learning structure in gene expression data using deep architectures, with an application to gene clustering. In: Bioinformatics and Biomedicine (BIBM), 2015 IEEE International Conference on. 2015. p. 1328–35.
Yuan Y, Shi Y, Li C, Kim J, Cai W, Han Z, et al. DeepGene : an advanced cancer type classifier based on deep learning and somatic point mutations. BMC Bioinformatics [Internet]. 2016;17(Suppl 17). Available from: http://dx.doi.org/10.1186/s12859-016-1334-9
Fawzy H, Kamel M, Al-amodi HSAB. Exploitation of Gene Expression and Cancer Biomarkers in Paving the Path to Era of Personalized Medicine. Genomics Proteomics Bioinformatics [Internet]. 2017;15(4):220–35. Available from: http://dx.doi.org/10.1016/j.gpb.2016.11.005
Lee Y, Lee C-K. Classification of multiple cancer types by multicategory support vector machines using gene expression data. Bioinformatics. 2003;19(9):1132–9.
Danaee P, Ghaeini R, Hendrix DA. A deep learning approach for cancer detection and relevant gene identification. In: PACIFIC SYMPOSIUM ON BIOCOMPUTING 2017. 2017. p. 219–29.
Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017;18(5):851–69.
Bhat RR, Viswanath V, Li X. DeepCancer : Detecting Cancer through Gene Expressions via Deep Generative Learning. (Ml).
Karabulut EM. Discriminative deep belief networks for microarray based cancer classification . 2017;28(3):1016–24.
Liu J, Wang X, Cheng Y, Zhang L. Tumor gene expression data classification via sample expansion- based deep learning. Oncotarget. 2017;8(65):109646.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57.
Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. 2002;1(March):203–9.
Rules C, Medjahed SA. Breast Cancer Diagnosis by using k-Nearest Neighbor with Different Breast Cancer Diagnosis by using k-Nearest Neighbor with Different Distances and Classification Rules. 2013;(January).
Mishra P, Bhoi N, Meher J. Effective clustering of microarray gene expression data using signal processing and soft computing methods. In: Electrical, Electronics, Signals, Communication and Optimization (EESCO), 2015 International Conference on. 2015. p. 1–4.
Woodward WA, Krishnamurthy S, Yamauchi H, El-Zein R, Ogura D, Kitadai E, et al. Genomic and expression analysis of microdissected inflammatory breast cancer. Breast Cancer Res Treat. 2013;138(3):761–72.
Kumar, C. ., & Muthumanickam, T. . (2023). Analysis of Unmanned Four-Wheeled Bot with AI Evaluation Feedback Linearization Method. International Journal on Recent and Innovation Trends in Computing and Communication, 11(2), 138–142. https://doi.org/10.17762/ijritcc.v11i2.6138
Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7(6):673.
Martinez-Ledesma E, Verhaak RGW, Treviño V. Identification of a multi-cancer gene expression biomarker for cancer clinical outcomes using a network-based algorithm. Sci Rep. 2015;5:11966.
Teixeira V, Camacho R, Ferreira PG. Learning influential genes on cancer gene expression data with stacked denoising autoencoders. In: Bioinformatics and Biomedicine (BIBM), 2017 IEEE International Conference on. 2017. p. 1201–5.
Microbe-host AI. ADAGE-Based Integration of Publicly Available Pseudomonas aeruginosa Gene Interactions. 1(1):1–17.
Braune E-B, Seshire A, Lendahl U. Notch and Wnt Dysregulation and Its Relevance for Breast Cancer and Tumor Initiation. Biomedicines. 2018;6(4):101. doi:10.3390/biomedicines6040101
Tammela T, Sanchez-Rivera FJ, Cetinbas NM, et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545(7654):355-359. doi:10.1038/nature22334
bin Saion, M. P. . (2021). Simulating Leakage Impact on Steel Industrial System Functionality. International Journal of New Practices in Management and Engineering, 10(03), 12–15. https://doi.org/10.17762/ijnpme.v10i03.129
Zhang M, Li H, Zou D, Gao J. Ruguo key genes and tumor driving factors identification of bladder cancer based on the RNA-seq profile. Onco Targets Ther. 2016;9:2717-2723. doi:10.2147/OTT.S92529
Ahmad I, Sansom OJ. Role of Wnt signalling in advanced prostate cancer. J Pathol. 2018;245(1):3-5. doi:10.1002/path.5029
Fakhar M, Najumuddin, Gul M, Rashid S. Antagonistic role of Klotho-derived peptides dynamics in the pancreatic cancer treatment through obstructing WNT-1 and Frizzled binding. Biophys Chem. 2018;240(June):107-117. doi:10.1016/j.bpc.2018.07.002
Fidalgo F, Rodrigues TC, Pinilla M, et al. Lymphovascular invasion and histologic grade are associated with specific genomic profiles in invasive carcinomas of the breast. Tumor Biol. 2015;36(3):1835-1848. doi:10.1007/s13277-014-2786-z
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Aditya Agnihotri, Mahesh Manchanda, Asif Ibrahim Tamboli

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





