Deep Learning Models for Cotton Leaf Disease Detection with VGG-16
Keywords:
Leaf Disease, Adam Optimizers, VGG-16, CNNAbstract
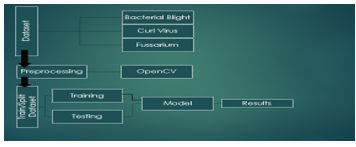
Cotton, potatoes, tomatoes, and chilies are the principal crops exported from Pakistan. Cotton is produced in greater quantities in India, China, and the US than in Pakistan. Eighty percent of all public oil is made up of cotton. Like other crops, it is being attacked by viruses, which lowers production and reduces economic revenue. There is misinformation on how illness diagnosis and care affect a region's ability to produce more. In this research, deep convolutional neural networks (CNN) were trained to recognize three forms of cotton leaf disease using a transform learning approach (Cotton leaf curl virus, fusarium wilt, bacterial blight). This study's main goal was to develop a single framework that could handle the challenging process of finding, identifying, and diagnosing cotton leaf disease. Additionally, to improve their performance on datasets of healthy and allergic cotton leaves, bigger weight parameter optimization using the Adam and RMSProp optimizers was explored. When compared to the other DL meta-architectures, Inception-VGG-16 trained with the feature extractor showed the greatest mean average accuracy. The recommended method was found to be new since it distinguished between leaf types that were healthy and those that were unhealthy. Using the DL approach to accurately identify cotton leaf disease would help to avoid the adverse effects of dietary management issues. On photos of cotton leaves, the trained model recognizes and labels the four classes (Cotton leaf curl virus, bacterial blight, fusarium wilt, healthy). CNN was 98% accurate overall.
Downloads
References
J. Zhi, T. Qiu, X. Bai, M. Xia, Z. Chen, and J. Zhou, "Effects of Nitrogen Conservation Measures on the Nitrogen Uptake by Cotton Plants and Nitrogen Residual in Soil Profile in Extremely Arid Areas of Xinjiang, China," Processes, vol. 10, no. 2, p. 353, 2022. [Online]. Available: https://www.mdpi.com/2227-9717/10/2/353.
T. Li, H. Zhao, S. Wang, C. Yang, and B. Huang, "Attack and Defense Strategy of Distribution Network Cyber-Physical System Considering EV Source-Charge Bidirectionality," Electronics, vol. 10, no. 23, p. 2973, 2021. [Online]. Available: https://www.mdpi.com/2079-9292/10/23/2973.
V. S. Dhaka et al., "A Survey of Deep Convolutional Neural Networks Applied for Prediction of Plant Leaf Diseases," Sensors, vol. 21, no. 14, p. 4749, 2021. [Online]. Available: https://www.mdpi.com/1424-8220/21/14/4749.
A. Arshad et al., "Impact of Climate Warming on Cotton Growth and Yields in China and Pakistan: A Regional Perspective," Agriculture, vol. 11, no. 2, p. 97, 2021. [Online]. Available: https://www.mdpi.com/2077-0472/11/2/97.
S. S. Zaidi et al., "Molecular insight into cotton leaf curl geminivirus disease resistance in cultivated cotton (Gossypium hirsutum)," (in eng), Plant Biotechnol J, vol. 18, no. 3, pp. 691-706, Mar 2020, doi: 10.1111/pbi.13236.
A. Rehman, A. A. Chandio, I. Hussain, and L. Jingdong, "Fertilizer consumption, water availability and credit distribution: Major factors affecting agricultural productivity in Pakistan," Journal of the Saudi Society of Agricultural Sciences, vol. 18, no. 3, pp. 269-274, 2019/07/01/ 2019, doi: https://doi.org/10.1016/j.jssas.2017.08.002.
A. Iqbal et al., "Genotypic Variation in Cotton Genotypes for Phosphorus-Use Efficiency," Agronomy, vol. 9, no. 11, p. 689, 2019. [Online]. Available: https://www.mdpi.com/2073-4395/9/11/689.
R. Koech and P. Langat, "Improving Irrigation Water Use Efficiency: A Review of Advances, Challenges and Opportunities in the Australian Context," Water, vol. 10, no. 12, p. 1771, 2018. [Online]. Available: https://www.mdpi.com/2073-4441/10/12/1771.
Z. Chen and D. R. Gallie, "The ascorbic acid redox state controls guard cell signaling and stomatal movement," (in eng), Plant Cell, vol. 16, no. 5, pp. 1143-62, May 2004, doi: 10.1105/tpc.021584.
D. R. Jones, "Plant Viruses Transmitted by Whiteflies," European Journal of Plant Pathology, vol. 109, no. 3, pp. 195-219, 2003/03/01 2003, doi: 10.1023/A:1022846630513.
M. Akila and P. Deepan, "Detection and Classification of Plant Leaf Diseases by using Deep Learning Algorithm," International journal of engineering research and technology, vol. 6, 2018.
R. W. Briddon, "Cotton leaf curl disease, a multicomponent begomovirus complex," Molecular Plant Pathology, vol. 4, no. 6, pp. 427-434, 2003, doi: https://doi.org/10.1046/j.1364-3703.2003.00188.x.
M. N. Sattar, Z. Iqbal, M. N. Tahir, and S. Ullah, "The Prediction of a New CLCuD Epidemic in the Old World," (in eng), Front Microbiol, vol. 8, p. 631, 2017, doi: 10.3389/fmicb.2017.00631.
J. Jenifer and D. J. B. Raja, "Skeleton Based Leaf Identification and Detection of Grape Plant Leaf Disease by Using K-Means Clustering Algorithm," 2017.
A. Ahmad et al., "Engineered Disease Resistance in Cotton Using RNA-Interference to Knock down Cotton leaf curl Kokhran virus-Burewala and Cotton leaf curl Multan betasatellite Expression," Viruses, vol. 9, no. 9, p. 257, 2017. [Online]. Available: https://www.mdpi.com/1999-4915/9/9/257.
K. L. Adams and J. F. Wendel, "Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids," (in eng), Genetics, vol. 171, no. 4, pp. 2139-42, Dec 2005, doi: 10.1534/genetics.105.047357.
L. M. Abualigah, A. T. Khader, M. A. Al-Betar, and O. A. Alomari, "Text feature selection with a robust weight scheme and dynamic dimension reduction to text document clustering," Expert Systems with Applications, vol. 84, pp. 24-36, 2017/10/30/ 2017, doi: https://doi.org/10.1016/j.eswa.2017.05.002.
V. S. Rana et al., "A Bemisia tabaci midgut protein interacts with begomoviruses and plays a role in virus transmission," (in eng), Cell Microbiol, vol. 18, no. 5, pp. 663-78, May 2016, doi: 10.1111/cmi.12538.
I. Korkontzelos, A. Nikfarjam, M. Shardlow, A. Sarker, S. Ananiadou, and G. H. Gonzalez, "Analysis of the effect of sentiment analysis on extracting adverse drug reactions from tweets and forum posts," (in eng), J Biomed Inform, vol. 62, pp. 148-58, Aug 2016, doi: 10.1016/j.jbi.2016.06.007.
P. R. Rothe and R. V. Kshirsagar, "Cotton leaf disease identification using pattern recognition techniques," 2015 International Conference on Pervasive Computing (ICPC), pp. 1-6, 2015.
D. Mondal, D. K. Kole, A. Chakraborty, and D. D. Majumder, "Detection and classification technique of Yellow Vein Mosaic Virus disease in okra leaf images using leaf vein extraction and Naive Bayesian classifier," 2015 International Conference on Soft Computing Techniques and Implementations (ICSCTI), pp. 166-171, 2015.
A. H. Paterson et al., "Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres," (in eng), Nature, vol. 492, no. 7429, pp. 423-7, Dec 20 2012, doi: 10.1038/nature11798.
M. Amlekar, A. T. Gaikwad, R. R. Manza, and P. L. Yannawar, "Leaf shape extraction for plant classification," 2015 International Conference on Pervasive Computing (ICPC), pp. 1-4, 2015.
M. S. Shahid, S. Mansoor, and R. W. Briddon, "Complete nucleotide sequences of cotton leaf curl Rajasthan virus and its associated DNA β molecule infecting tomato," Archives of Virology, vol. 152, no. 11, pp. 2131-2134, 2007/10/01 2007, doi: 10.1007/s00705-007-1043-9.
S. S. Sohrab, E. I. Azhar, M. A. Kamal, P. S. Bhattacharya, and D. Rana, "Genetic variability of Cotton leaf curl betasatellite in Northern India," Saudi Journal of Biological Sciences, vol. 21, no. 6, pp. 626-631, 2014/12/01/ 2014, doi: https://doi.org/10.1016/j.sjbs.2014.11.006.
R. Guy, "Water-use efficiency and productivity trends in Australian irrigated cotton: a review," (in English), Crop & pasture science, vol. v. 64, no. no. 12, pp. pp. 1033-1048-2013 v.64 no.12, 0000 2013, doi: 10.1071/CP13315.
Z. Jian and Z. Wei, "Support vector machine for recognition of cucumber leaf diseases," 2010 2nd International Conference on Advanced Computer Control, vol. 5, pp. 264-266, 2010.
W. Nazeer, A. L. Tipu, S. Ahmad, K. Mahmood, A. Mahmood, and B. Zhou, "Evaluation of cotton leaf curl virus resistance in BC1, BC2, and BC3 progenies from an interspecific cross between Gossypium arboreum and Gossypium hirsutum," (in eng), PLoS One, vol. 9, no. 11, p. e111861, 2014, doi: 10.1371/journal.pone.0111861.
W. Malik et al., "Molecular markers and cotton genetic improvement: current status and future prospects," (in eng), ScientificWorldJournal, vol. 2014, p. 607091, 2014, doi: 10.1155/2014/607091.
S. Akhtar et al., "Regional Changes in the Sequence of Cotton Leaf Curl Multan Betasatellite," Viruses, vol. 6, no. 5, pp. 2186-2203, 2014. [Online]. Available: https://www.mdpi.com/1999-4915/6/5/2186.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





