Data Embeddings in Medical Applications: A Survey of Techniques and Applications
Keywords:
data embeddings, medical data, principal component analysis, t-distributed stochastic embedding, autoencodersAbstract
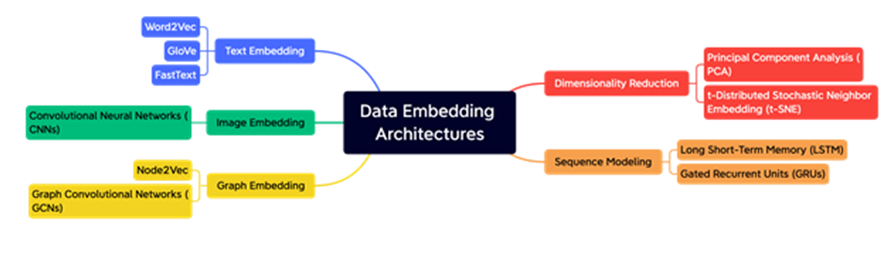
The abundance of medical data available, ranging from electronic health records to medical images, presents a unique opportunity to gain valuable insights into disease processes, improve treatments, and enhance patient outcomes. However, the complexity, high dimensionality, and heterogeneity of these datasets pose significant challenges to their analysis and interpretation. One technique that has gained popularity for addressing these challenges is data embeddings. Data embeddings are low-dimensional representations of high-dimensional data that preserve the underlying structure and relationships between data points. In the medical domain, data embeddings have found numerous applications, such as disease diagnosis, patient risk stratification, and drug discovery. This survey paper aims to provide a comprehensive overview of data embeddings techniques and their applications in the medical domain. The paper introduces the concept of data embeddings and their properties, and provides a detailed discussion of popular embedding techniques, including principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), and autoencoders. The paper also reviews various applications of data embeddings in the medical domain, such as disease diagnosis, patient clustering, and drug discovery. The paper concludes with a discussion of future directions and emerging trends in data embeddings for medical applications, emphasizing the need for more robust and interpretable embedding techniques and the importance of considering clinical context when developing and applying these techniques.
Downloads
References
Keidar, D., Yaron, D., Goldstein, E., Shachar, Y., Blass, A., Charbinsky, L., Aharony, I., Lifshitz, L., Lumelsky, D., Neeman, Z., Mizrachi, M., Hajouj, M., Eizenbach, N., Sela, E., Weiss, C. S., Levin, P., Benjaminov, O., Bachar, G. N., Tamir, S., … Eldar, Y. C. (2021). COVID-19 classification of X-ray images using deep neural networks. European Radiology, 31(12), 9654–9663. https://doi.org/10.1007/s00330-021-08050-1
Despotovic, V., Ismael, M., Cornil, M., Call, R. M., & Fagherazzi, G. (2021). Detection of COVID-19 from voice, cough and breathing patterns: Dataset and preliminary results. Computers in Biology and Medicine, 138(104944), 104944. https://doi.org/10.1016/j.compbiomed.2021.104944
Kumar, V., Recupero, D. R., Riboni, D., & Helaoui, R. (2021). Ensembling Classical Machine Learning and Deep Learning Approaches for Morbidity Identification from Clinical Notes. IEEE Access, 9, 7107–7126. https://doi.org/10.1109/ACCESS.2020.3043221
Ye, J., Yao, L., Shen, J., Janarthanam, R., & Luo, Y. (2020). Predicting mortality in critically ill patients with diabetes using machine learning and clinical notes. BMC Medical Informatics and Decision Making, 20(Suppl 11), 295. https://doi.org/10.1186/s12911-020-01318-4
Heo, T. S., Kim, Y. S., Choi, J. M., Jeong, Y. S., Seo, S. Y., Lee, J. H., Jeon, J. P., & Kim, C. (2020). Prediction of stroke outcome using natural language processing-based machine learning of radiology report of brain MRI. Journal of Personalized Medicine, 10(4), 1–11. https://doi.org/10.3390/jpm10040286
Tian, Y., Shen, W., Song, Y., Xia, F., He, M., & Li, K. (2020). Improving biomedical named entity recognition with syntactic information. BMC Bioinformatics, 21(1), 539. https://doi.org/10.1186/s12859-020-03834-6
Landi, I., Glicksberg, B. S., Lee, H. C., Cherng, S., Landi, G., Danieletto, M., Dudley, J. T., Furlanello, C., & Miotto, R. (2020). Deep representation learning of electronic health records to unlock patient stratification at scale. Npj Digital Medicine, 3(1), 96. https://doi.org/10.1038/s41746-020-0301-z
Wang, H., Li, Y., Khan, S. A., & Luo, Y. (2020). Prediction of breast cancer distant recurrence using natural language processing and knowledge-guided convolutional neural network. Artificial Intelligence in Medicine, 110(101977), 101977. https://doi.org/10.1016/j.artmed.2020.101977
Luo, J. W., & Chong, J. J. R. (2020). Review of Natural Language Processing in Radiology. Neuroimaging Clinics of North America, 30(4), 447–458. https://doi.org/10.1016/j.nic.2020.08.001
Afzal, M., Alam, F., Malik, K. M., & Malik, G. M. (2020). Clinical Context–Aware Biomedical Text Summarization Using Deep Neural Network: Model Development and Validation. Journal of Medical Internet Research, 22(10), e19810. https://doi.org/10.2196/19810
Barash, Y., Guralnik, G., Tau, N., Soffer, S., Levy, T., Shimon, O., Zimlichman, E., Konen, E., & Klang, E. (2020). Comparison of deep learning models for natural language processing-based classification of non-English head CT reports. Neuroradiology, 62(10), 1247–1256. https://doi.org/10.1007/s00234-020-02420-0
Obeid, J. S., Davis, M., Turner, M., Meystre, S. M., Heider, P. M., O’Bryan, E. C., & Lenert, L. A. (2020). An artificial intelligence approach to COVID-19 infection risk assessment in virtual visits: A case report. Journal of the American Medical Informatics Association, 27(8), 1321–1325. https://doi.org/10.1093/jamia/ocaa105
Chen, B., Li, J., Lu, G., Yu, H., & Zhang, D. (2020). Label co-occurrence learning with graph convolutional networks for multi-label chest X-ray image classification. IEEE Journal of Biomedical and Health Informatics, 24(8), 2292–2302. https://doi.org/10.1109/JBHI.2020.2967084
Chen, C. H., Hsieh, J. G., Cheng, S. L., Lin, Y. L., Lin, P. H., & Jeng, J. H. (2020). Emergency department disposition prediction using a deep neural network with integrated clinical narratives and structured data. International Journal of Medical Informatics, 139(104146), 104146. https://doi.org/10.1016/j.ijmedinf.2020.104146
Blanco, A., Perez-de-Viñaspre, O., Pérez, A., & Casillas, A. (2020). Boosting ICD multi-label classification of health records with contextual embeddings and label-granularity. Computer Methods and Programs in Biomedicine, 188(105264), 105264. https://doi.org/10.1016/j.cmpb.2019.105264
Malik, K. M., Krishnamurthy, M., Alobaidi, M., Hussain, M., Alam, F., & Malik, G. (2020). Automated domain-specific healthcare knowledge graph curation framework: Subarachnoid hemorrhage as phenotype. Expert Systems with Applications, 145(113120), 113120. https://doi.org/10.1016/j.eswa.2019.113120
Korach, Z. T., Yang, J., Rossetti, S. C., Cato, K. D., Kang, M. J., Knaplund, C., Schnock, K. O., Garcia, J. P., Jia, H., Schwartz, J. M., & Zhou, L. (2020). Mining clinical phrases from nursing notes to discover risk factors of patient deterioration. International Journal of Medical Informatics, 135(104053), 104053. https://doi.org/10.1016/j.ijmedinf.2019.104053
Akhtyamova, L., Martínez, P., Verspoor, K., & Cardiff, J. (2020). Testing contextualized word embeddings to improve NER in Spanish clinical case narratives. IEEE Access, 8, 164717–164726. https://doi.org/10.1109/ACCESS.2020.3018688
Dai, H. J., Su, C. H., & Wu, C. S. (2020). Adverse drug event and medication extraction in electronic health records via a cascading architecture with different sequence labeling models and word embeddings. Journal of the American Medical Informatics Association, 27(1), 47–55. https://doi.org/10.1093/jamia/ocz120
Dai, H. J., Su, C. H., & Wu, C. S. (2020). Adverse drug event and medication extraction in electronic health records via a cascading architecture with different sequence labeling models and word embeddings. Journal of the American Medical Informatics Association, 27(1), 47–55. https://doi.org/10.1093/jamia/ocz120
Gligic, L., Kormilitzin, A., Goldberg, P., & Nevado-Holgado, A. (2020). Named entity recognition in electronic health records using transfer learning bootstrapped Neural Networks. Neural Networks, 121, 132–139. https://doi.org/10.1016/j.neunet.2019.08.032
Yang, X., Lyu, T., Li, Q., Lee, C. Y., Bian, J., Hogan, W. R., & Wu, Y. (2019). A study of deep learning methods for de-identification of clinical notes in cross-institute settings. BMC Medical Informatics and Decision Making, 19(Suppl 5), 232. https://doi.org/10.1186/s12911-019-0935-4
Yang, X., Lyu, T., Li, Q., Lee, C. Y., Bian, J., Hogan, W. R., & Wu, Y. (2019). A study of deep learning methods for de-identification of clinical notes in cross-institute settings. BMC Medical Informatics and Decision Making, 19(Suppl 5), 232. https://doi.org/10.1186/s12911-019-0935-4
Gao, C., Osmundson, S., Velez Edwards, D. R., Jackson, G. P., Malin, B. A., & Chen, Y. (2019). Deep learning predicts extreme preterm birth from electronic health records. Journal of Biomedical Informatics, 100(103334), 103334. https://doi.org/10.1016/j.jbi.2019.103334
Topaz, M., Murga, L., Bar-Bachar, O., McDonald, M., & Bowles, K. (2019). NimbleMiner: An Open-Source Nursing-Sensitive Natural Language Processing System Based on Word Embedding. CIN - Computers Informatics Nursing, 37(11), 583–590. https://doi.org/10.1097/CIN.0000000000000557
Zhang, Y., Lin, H., Yang, Z., Wang, J., Sun, Y., Xu, B., & Zhao, Z. (2019). Neural network-based approaches for biomedical relation classification: A review. Journal of Biomedical Informatics, 99(103294), 103294. https://doi.org/10.1016/j.jbi.2019.103294
Suárez-Paniagua, V., Rivera Zavala, R. M., Segura-Bedmar, I., & Martínez, P. (2019). A two-stage deep learning approach for extracting entities and relationships from medical texts. Journal of Biomedical Informatics, 99(103285), 103285. https://doi.org/10.1016/j.jbi.2019.103285
Batbaatar, E., & Ryu, K. H. (2019). Ontology-based healthcare named entity recognition from twitter messages using a recurrent neural network approach. International Journal of Environmental Research and Public Health, 16(19), 3628. https://doi.org/10.3390/ijerph16193628
Dai, H. J., & Wang, C. K. (2019). Classifying adverse drug reactions from imbalanced twitter data. International Journal of Medical Informatics, 129, 122–132. https://doi.org/10.1016/j.ijmedinf.2019.05.017
Trivedi, G., Hong, C., Dadashzadeh, E. R., Handzel, R. M., Hochheiser, H., & Visweswaran, S. (2019). Identifying incidental findings from radiology reports of trauma patients: An evaluation of automated feature representation methods. International Journal of Medical Informatics, 129, 81–87. https://doi.org/10.1016/j.ijmedinf.2019.05.021
Obeid, J. S., Weeda, E. R., Matuskowitz, A. J., Gagnon, K., Crawford, T., Carr, C. M., & Frey, L. J. (2019). Automated detection of altered mental status in emergency department clinical notes: A deep learning approach. BMC Medical Informatics and Decision Making, 19(1), 164. https://doi.org/10.1186/s12911-019-0894-9
Yang, Y., Wang, X., Huang, Y., Chen, N., Shi, J., & Chen, T. (2019). Ontology-based venous thromboembolism risk assessment model developing from medical records. BMC Medical Informatics and Decision Making, 19(Suppl 4), 151. https://doi.org/10.1186/s12911-019-0856-2
Oleynik, M., Kugic, A., Kasáč, Z., & Kreuzthaler, M. (2019). Evaluating shallow and deep learning strategies for the 2018 n2c2 shared task on clinical text classification. Journal of the American Medical Informatics Association, 26(11), 1247–1254. https://doi.org/10.1093/jamia/ocz149
Si, Y., Wang, J., Xu, H., & Roberts, K. (2019). Enhancing clinical concept extraction with contextual embeddings. Journal of the American Medical Informatics Association, 26(11), 1297–1304. https://doi.org/10.1093/jamia/ocz096
Gargiulo, F., Silvestri, S., Ciampi, M., & De Pietro, G. (2019). Deep neural network for hierarchical extreme multi-label text classification. Applied Soft Computing Journal, 79, 125–138. https://doi.org/10.1016/j.asoc.2019.03.041
Yao, L., Mao, C., & Luo, Y. (2019). Clinical text classification with rule-based features and knowledge-guided convolutional neural networks. BMC Medical Informatics and Decision Making, 19(Suppl 3), 71. https://doi.org/10.1186/s12911-019-0781-4
Sheikhalishahi, S., Miotto, R., Dudley, J. T., Lavelli, A., Rinaldi, F., & Osmani, V. (2019). Natural language processing of clinical notes on chronic diseases: Systematic review. JMIR Medical Informatics, 7(2), e12239. https://doi.org/10.2196/12239
Dimitriadis, D., & Tsoumakas, G. (2019). Word embeddings and external resources for answer processing in biomedical factoid question answering. Journal of Biomedical Informatics, 92(103118), 103118. https://doi.org/10.1016/j.jbi.2019.103118
Ning, W., Chan, S., Beam, A., Yu, M., Geva, A., Liao, K., Mullen, M., Mandl, K. D., Kohane, I., Cai, T., & Yu, S. (2019). Feature extraction for phenotyping from semantic and knowledge resources. Journal of Biomedical Informatics, 91(103122), 103122. https://doi.org/10.1016/j.jbi.2019.103122
Li, Y., Jin, R., & Luo, Y. (2019). Classifying relations in clinical narratives using segment graph convolutional and recurrent neural networks (Seg-GCRNs). Journal of the American Medical Informatics Association, 26(3), 262–268. https://doi.org/10.1093/jamia/ocy157
Li, X., Wang, H., He, H., Du, J., Chen, J., & Wu, J. (2019). Intelligent diagnosis with Chinese electronic medical records based on convolutional neural networks. BMC Bioinformatics, 20(1), 62. https://doi.org/10.1186/s12859-019-2617-8
Topaz, M., Murga, L., Gaddis, K. M., McDonald, M. V., Bar-Bachar, O., Goldberg, Y., & Bowles, K. H. (2019). Mining fall-related information in clinical notes: Comparison of rule-based and novel word embedding-based machine learning approaches. Journal of Biomedical Informatics, 90(103103), 103103. https://doi.org/10.1016/j.jbi.2019.103103
Wunnava, S., Qin, X., Kakar, T., Sen, C., Rundensteiner, E. A., & Kong, X. (2019). Adverse Drug Event Detection from Electronic Health Records Using Hierarchical Recurrent Neural Networks with Dual-Level Embedding. Drug Safety, 42(1), 113–122. https://doi.org/10.1007/s40264-018-0765-9
Chapman, A. B., Peterson, K. S., Alba, P. R., DuVall, S. L., & Patterson, O. V. (2019). Detecting Adverse Drug Events with Rapidly Trained Classification Models. Drug Safety, 42(1), 147–156. https://doi.org/10.1007/s40264-018-0763-y
Guan, M., Cho, S., Petro, R., Zhang, W., Pasche, B., & Topaloglu, U. (2019). Natural language processing and recurrent network models for identifying genomic mutation-associated cancer treatment change from patient progress notes. JAMIA Open, 2(1), 139–149. https://doi.org/10.1093/jamiaopen/ooy061
Zhao, B. (2019). Clinical Data Extraction and Normalization of Cyrillic Electronic Health Records Via Deep-Learning Natural Language Processing. JCO Clinical Cancer Informatics, 3(3), 1–9. https://doi.org/10.1200/cci.19.00057
Fan, Y., Pakhomov, S., McEwan, R., Zhao, W., Lindemann, E., & Zhang, R. (2019). Using word embeddings to expand terminology of dietary supplements on clinical notes. JAMIA Open, 2(2), 246–253. https://doi.org/10.1093/jamiaopen/ooz007
Chowdhury, S., Dong, X., Qian, L., Li, X., Guan, Y., Yang, J., & Yu, Q. (2018). A multitask bi-directional RNN model for named entity recognition on Chinese electronic medical records. BMC Bioinformatics, 19(17), 75–84. https://doi.org/10.1186/s12859-018-2467-9
Xu, K., Zhou, Z., Gong, T., Hao, T., & Liu, W. (2018). SBLC: A hybrid model for disease named entity recognition based on semantic bidirectional LSTMs and conditional random fields. BMC Medical Informatics and Decision Making, 18(Suppl 5), 114. https://doi.org/10.1186/s12911-018-0690-y
Catling, F., Spithourakis, G. P., & Riedel, S. (2018). Towards automated clinical coding. International Journal of Medical Informatics, 120, 50–61. https://doi.org/10.1016/j.ijmedinf.2018.09.021
Song, H. J., Jo, B. C., Park, C. Y., Kim, J. D., & Kim, Y. S. (2018). Comparison of named entity recognition methodologies in biomedical documents. BioMedical Engineering Online, 17(2), 1–14. https://doi.org/10.1186/s12938-018-0573-6
Segura-Bedmar, I., Colón-Ruíz, C., Tejedor-Alonso, M. Á., & Moro-Moro, M. (2018). Predicting of anaphylaxis in big data EMR by exploring machine learning approaches. Journal of Biomedical Informatics, 87, 50–59. https://doi.org/10.1016/j.jbi.2018.09.012
Wang, Y., Liu, S., Afzal, N., Rastegar-Mojarad, M., Wang, L., Shen, F., Kingsbury, P., & Liu, H. (2018). A comparison of word embeddings for the biomedical natural language processing. Journal of Biomedical Informatics, 87, 12–20. https://doi.org/10.1016/j.jbi.2018.09.008
Ye, C., & Fabbri, D. (2018). Extracting similar terms from multiple EMR-based semantic embeddings to support chart reviews. Journal of Biomedical Informatics, 83, 63–72. https://doi.org/10.1016/j.jbi.2018.05.014
Menger, V., Scheepers, F., & Spruit, M. (2018). Comparing deep learning and classical machine learning approaches for predicting inpatient violence incidents from clinical text. Applied Sciences (Switzerland), 8(6), 981. https://doi.org/10.3390/app8060981
Zech, J., Pain, M., Titano, J., Badgeley, M., Schefflein, J., Su, A., Costa, A., Bederson, J., Lehar, J., & Oermann, E. K. (2018). Natural language-based machine learning models for the annotation of clinical radiology reports. Radiology, 287(2), 570–580. https://doi.org/10.1148/radiol.2018171093
Zhou, D., Miao, L., & He, Y. (2018). Position-aware deep multi-task learning for drug–drug interaction extraction. Artificial Intelligence in Medicine, 87, 1–8. https://doi.org/10.1016/j.artmed.2018.03.001
Koola, J. D., Davis, S. E., Al-Nimri, O., Parr, S. K., Fabbri, D., Malin, B. A., Ho, S. B., & Matheny, M. E. (2018). Development of an automated phenotyping algorithm for hepatorenal syndrome. Journal of Biomedical Informatics, 80, 87–95. https://doi.org/10.1016/j.jbi.2018.03.001
Jadhav, S. B. ., & Kodavade, D. V. . (2023). Enhancing Flight Delay Prediction through Feature Engineering in Machine Learning Classifiers: A Real Time Data Streams Case Study. International Journal on Recent and Innovation Trends in Computing and Communication, 11(2s), 212–218. https://doi.org/10.17762/ijritcc.v11i2s.6064
Duarte, F., Martins, B., Pinto, C. S., & Silva, M. J. (2018). Deep neural models for ICD-10 coding of death certificates and autopsy reports in free-text. Journal of Biomedical Informatics, 80, 64–77. https://doi.org/10.1016/j.jbi.2018.02.011
Si, Y., & Roberts, K. (2018). A Frame-Based NLP System for Cancer-Related Information Extraction. AMIA ... Annual Symposium Proceedings. AMIA Symposium, 2018, 1524–1533. /pmc/articles/PMC6371330/
Glicksberg, B. S., Miotto, R., Johnson, K. W., Shameer, K., Li, L., Chen, R., & Dudley, J. T. (2018). Automated disease cohort selection using word embeddings from Electronic Health Records. Pacific Symposium on Biocomputing, 0(212669), 145–156. https://doi.org/10.1142/9789813235533_0014
Luo, Y., Cheng, Y., Uzuner, Ö., Szolovits, P., & Starren, J. (2018). Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. Journal of the American Medical Informatics Association, 25(1), 93–98. https://doi.org/10.1093/jamia/ocx090
Xie, J., Liu, X., & Zeng, D. D. (2018). Mining e-cigarette adverse events in social media using Bi-LSTM recurrent neural network with word embedding representation. Journal of the American Medical Informatics Association, 25(1), 72–80. https://doi.org/10.1093/jamia/ocx045
Banerjee, I., Chen, M. C., Lungren, M. P., & Rubin, D. L. (2018). Radiology report annotation using intelligent word embeddings: Applied to multi-institutional chest CT cohort. Journal of Biomedical Informatics, 77, 11–20. https://doi.org/10.1016/j.jbi.2017.11.012
Wang, A., Wang, J., Lin, H., Zhang, J., Yang, Z., & Xu, K. (2017). A multiple distributed representation method based on neural network for biomedical event extraction. BMC Medical Informatics and Decision Making, 17(Suppl 3). https://doi.org/10.1186/s12911-017-0563-9
Weng, W. H., Wagholikar, K. B., McCray, A. T., Szolovits, P., & Chueh, H. C. (2017). Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach. BMC Medical Informatics and Decision Making, 17(1). https://doi.org/10.1186/s12911-017-0556-8
Jauregi Unanue, I., Zare Borzeshi, E., & Piccardi, M. (2017). Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition. Journal of Biomedical Informatics, 76, 102–109. https://doi.org/10.1016/j.jbi.2017.11.007
Lin, C., Hsu, C. J., Lou, Y. S., Yeh, S. J., Lee, C. C., Su, S. L., & Chen, H. C. (2017). Artificial intelligence learning semantics via external resources for classifying diagnosis codes in discharge notes. Journal of Medical Internet Research, 19(11). https://doi.org/10.2196/jmir.8344
Lyu, C., Chen, B., Ren, Y., & Ji, D. (2017). Long short-term memory RNN for biomedical named entity recognition. BMC Bioinformatics, 18(1), 462. https://doi.org/10.1186/s12859-017-1868-5
Cho, H., Choi, W., & Lee, H. (2017). A method for named entity normalization in biomedical articles: Application to diseases and plants. BMC Bioinformatics, 18(1). https://doi.org/10.1186/s12859-017-1857-8
Sulieman, L., Gilmore, D., French, C., Cronin, R. M., Jackson, G. P., Russell, M., & Fabbri, D. (2017). Classifying patient portal messages using Convolutional Neural Networks. Journal of Biomedical Informatics, 74, 59–70. https://doi.org/10.1016/j.jbi.2017.08.014
Mr. Anish Dhabliya. (2013). Ultra Wide Band Pulse Generation Using Advanced Design System Software . International Journal of New Practices in Management and Engineering, 2(02), 01 - 07. Retrieved from http://ijnpme.org/index.php/IJNPME/article/view/14
Jimeno Yepes, A. (2017). Word embeddings and recurrent neural networks based on Long-Short Term Memory nodes in supervised biomedical word sense disambiguation. Journal of Biomedical Informatics, 73, 137–147. https://doi.org/10.1016/j.jbi.2017.08.001
Kuang, S., & Davison, B. D. (2017). Learning word embeddings with chi-square weights for healthcare tweet classification. Applied Sciences (Switzerland), 7(8), 846. https://doi.org/10.3390/app7080846
Luo, Y. (2017). Recurrent neural networks for classifying relations in clinical notes. Journal of Biomedical Informatics, 72, 85–95. https://doi.org/10.1016/j.jbi.2017.07.006
Tao, C., Filannino, M., & Uzuner, Ö. (2017). Prescription extraction using CRFs and word embeddings. Journal of Biomedical Informatics, 72, 60–66. https://doi.org/10.1016/j.jbi.2017.07.002
Gridach, M. (2017). Character-level neural network for biomedical named entity recognition. Journal of Biomedical Informatics, 70, 85–91. https://doi.org/10.1016/j.jbi.2017.05.002
Nguyen, T., Larsen, M. E., O’Dea, B., Phung, D., Venkatesh, S., & Christensen, H. (2017). Estimation of the prevalence of adverse drug reactions from social media. International Journal of Medical Informatics, 102, 130–137. https://doi.org/10.1016/j.ijmedinf.2017.03.013
Kang, T., Zhang, S., Xu, N., Wen, D., Zhang, X., & Lei, J. (2017). Detecting negation and scope in Chinese clinical notes using character and word embedding. Computer Methods and Programs in Biomedicine, 140, 53–59. https://doi.org/10.1016/j.cmpb.2016.11.009
Wu, Y., Jiang, M., Xu, J., Zhi, D., & Xu, H. (2017). Clinical Named Entity Recognition Using Deep Learning Models. AMIA ... Annual Symposium Proceedings. AMIA Symposium, 2017, 1812–1819.
Banerjee, I., Madhavan, S., Goldman, R. E., & Rubin, D. L. (2017). Intelligent Word Embeddings of Free-Text Radiology Reports. AMIA ... Annual Symposium Proceedings. AMIA Symposium, 2017, 411–420. https://arxiv.org/abs/1711.06968v1
Zhang, S., Kang, T., Zhang, X., Wen, D., Elhadad, N., & Lei, J. (2016). Speculation detection for Chinese clinical notes: Impacts of word segmentation and embedding models. Journal of Biomedical Informatics, 60, 334–341. https://doi.org/10.1016/j.jbi.2016.02.011
Chen, C., Zhao, X., Wang, J., Li, D., Guan, Y., & Hong, J. (2022). Dynamic graph convolutional network for assembly behavior recognition based on attention mechanism and multi-scale feature fusion. Scientific Reports, 12(1), 1–13. https://doi.org/10.1038/s41598-022-11206-8
Raza, A., Mehmood, A., Ullah, S., Ahmad, M., Choi, G. S., & On, B. W. (2019). Heartbeat sound signal classification using deep learning. Sensors (Switzerland), 19(21). https://doi.org/10.3390/s19214819
Varghese, A., Agyeman-Badu, G., & Cawley, M. (2020). Deep learning in automated text classification: a case study using toxicological abstracts. Environment Systems and Decisions, 40(4), 465–479. https://doi.org/10.1007/s10669-020-09763-2
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
All papers should be submitted electronically. All submitted manuscripts must be original work that is not under submission at another journal or under consideration for publication in another form, such as a monograph or chapter of a book. Authors of submitted papers are obligated not to submit their paper for publication elsewhere until an editorial decision is rendered on their submission. Further, authors of accepted papers are prohibited from publishing the results in other publications that appear before the paper is published in the Journal unless they receive approval for doing so from the Editor-In-Chief.
IJISAE open access articles are licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. This license lets the audience to give appropriate credit, provide a link to the license, and indicate if changes were made and if they remix, transform, or build upon the material, they must distribute contributions under the same license as the original.





